Abstract
Human retinoblastoma occurs in two forms (familial and sporadic) both due to biallelic mutation of the RB1/p105 gene even if its loss is insufficient for malignancy. We have recently reported that loss of expression of the retinoblastoma-related protein pRb2/p130 correlates with low apoptotic index, suggesting that RB2/p130 gene could be involved in retinoblastoma. Mutational analysis of RB2/p130 in primary tumors showed a tight correlation between Exon 1 mutations and pRb2/p130 expression level in sporadic retinoblastoma. These mutations are located within a CpG-enriched region prone to de novo methylation. Analysis of RB2/p130 methylation status revealed that epigenetic events, most probably consequent to the Exon 1 mutations, determined the observed phenotype. Treatment of Weri-Rb1 cell line by 5-Aza-dC induced an increase in expression level of pRb2/p130, E2F1, p73 and p53. Overall, our results highlight a crucial role of epigenetic events in sporadic retinoblastoma, which opens a perspective for new therapeutic approaches.
Similar content being viewed by others
Introduction
On the path to becoming cancerous, normal mammalian cells evolve to an atypical organization. Normal tissues and cells are the product of a complex set of genes expressed at different levels, which respond to internal and external stimuli to alter level of expression. Cancer cells do not express the same set of genes as their normal counterparts, and such anarchy is a hallmark of the cancerous state (Hanahan and Weinberg, 2000). The change in gene expression profiles evident in cancer cells depends on alterations at the genetic (i.e. mutations) as well as epigenetic (i.e. transcriptional inactivation due to methylation) level (Issa, 1999; Knudson, 2002). Retinoblastoma is the most common intraocular malignancy in children. Human retinoblastoma occurs in two forms: a nonheritable form, which is usually unilateral, and a heritable form, which is often bilateral with autosomal dominant expression. Both forms have been ascribed to biallelic mutation of the Rb1/p105 gene and the consequent loss of its tumor-suppressive functions. In the familial form, one mutated copy is inherited from an affected parent and a further mutation accumulates to determine the disease, while in the sporadic form both mutations occur spontaneously (Knudson et al., 1975). The basic function of pRb1/p105 is to hold cells in G1 or G0 phase of the cell cycle and prevent entry into S phase by interacting and negatively regulating the E2F family of transcription factors. Moreover, pRb1/p105 is also involved in the apoptotic response by interacting with p53 pro-apoptotic pathway (Hsieh et al., 1997, 1999; Chan et al., 2001). Not withstanding the fact that mutation of Rb1/p105 is common to all retinoblastomas, much evidence indicates that loss of pRb1/p105 from a developing retinal cell is insufficient for malignancy (DiCiommo et al., 2000). The possibility that further mutations of other tumor suppressor genes could occur in sporadic retinoblastoma is supported by recent studies providing direct evidence that loss of pRb1/p105 function leads to genome instability and predispose to cancer by increasing DNA mutation rate (Zheng and Lee, 2002). On the other hand, in tumorigenesis, genomic instability is tightly linked to deregulation of DNA methylation pattern and partial or total gene silencing may be achieved by this epigenetic mechanism (Sugimura and Ushijima, 2000). This is the case also for retinoblastoma where hypermethylation of both RB1/p105 and other genes has been shown (Choy et al., 2004). pRb1/p105 functions are shared by two homologous proteins, pRb2/p130 and p107, so that the three of them are referred to as retinoblastoma family proteins (pRBs) (Cinti and Giordano, 2000). In particular, RB2/p130 gene has been found mutated or functionally inactivated in many tumors and its role in controlling p53-independent apoptotic response has been elucidated recently (La Sala et al., 2003). Interestingly, we reported recently (Bellan et al., 2002) that, in some sporadic retinoblastomas, also the expression of pRb2/p130 is impaired and its loss of expression correlates with low apoptotic index, suggesting that in sporadic retinoblastoma also RB2/p130 gene could be mutated or functionally inactivated.
Therefore, we performed RB2/p130 mutational analysis in familial and sporadic retinoblastoma in an attempt to correlate pRb2/p130 expression level with gene mutational status. Moreover, we analysed RB2/p130 promoter methylation pattern to investigate the relative weight of genetic and epigenetic events in determining the alteration of pRb2/p130 expression level.
Results
Significance of pRb2 / p130 expression level with respect to mutational status
Experiments were performed on 10 retinoblastoma primary tumors (two familial and eight sporadic), Weri-Rb1 retinoblastoma cell line and three normal retina samples. pRb1/p105 and pRb2/p130 expression levels were evaluated by Western blot analysis in normal retina (NR), in three frozen sporadic retinoblastoma samples (samples 8–10) and in Weri-Rb1 cell line (W). Data are shown in Figure 1a where the lack of pRb1/p105 expression and the downregulation of pRb2/p130 in both primary tumors and Weri-Rb1 cells is evident. pRb2/p130 downregulation was confirmed on sections from paraffin-embedded tissues by immunohistochemical analysis, which has been extended to seven more retinoblastoma cases (Figure 1b and c). This further analysis evidenced differences in pRb2/p130 expression level among various patients (Table 1). In particular, downregulation was detected in four out of 10 samples, while in four cases pRb2/p130 was not expressed and in the remaining two cases there was no difference with respect to normal retina. The only correlation between this result and clinicopathological classification was that both samples indistinguishable from normal retina arise from bilateral familial retinoblastoma (B/F) patients. To check whether differences in pRb2/p130 expression level depend on different mutational patterns, we screened DNA samples for mutations in RB2/p130 coding regions using properly designed primers able to amplify each of the 22 exons (Table 2a). Tissue from normal retinas and tumor specimens were isolated by laser capture microdissection (Figure 1d–f), DNA was extracted and processed by PCR. The PCR-product sequences have been matched with the wild-type sequences (Gene Bank Accession Numbers X74594 and U53220) (Mayol et al., 1993; Baldi et al., 1996). With the exception of one familial retinoblastoma patient, we detected two Exon 1 mutations at nucleotides 178 and 259 (TCT → CCT: SER → PRO and CCC → GCC: PRO → ALA) in all primary tumors (Table 1, Figure 1g). Exon 1 mutations were not detected either in normal retina samples or in 15 healthy donors' blood samples (Table 1). In primary tumors, we found a clear correlation between pRb2/p130 expression level and exon 1 homozygous/heterozygous mutations. In particular, loss of expression is correlated with double homozygous mutation (Table 1, samples 3–5, 9), and weak expression coincides with the presence of 178 homozygous and 259 heterozygous mutations (Table 1, samples 6–8, 10), while when both mutations are heterozygous, the expression level is normal (Table 1, sample 2). Screening of RB2/p130 mutational pattern in Weri-Rb1 retinoblastoma cell line evidenced the same Exon 1 homozygous mutations observed in nine out of 10 primary tumors. Further mutations were detected in both primary tumor and Weri-Rb1 retinoblastoma cell line in Exons 4, 6, 13, 16 and 21 (see Table 1, for details). We have also found three additional homozygous silent mutations in Exons 15 and 17 present in both control (normal retina and healthy donors) and retinoblastoma samples (primary tumors and Weri-Rb1 cell line). A further homozygous silent mutation in Exon 12 was detectable only in sporadic retinoblastoma tumors (six out of eight samples) (Table 1). These results suggest that the two silent mutations in Exons 15 and 17 may represent a gene polymorphism occurring naturally in the population, while the homozygous silent mutation in Exon 12 may be specific of sporadic retinoblastoma and therefore predictive with respect to tumor phenotype.
pRb1/p105 and pRb2/p130 protein expression levels and mutational analysis in human cell lines and in tumor samples. (a) Western Blot analysis of pRb1/p105 and pRb2/p130 in Jurkat cell line used as positive control for antibodies (C), normal retina (NR), Weri-Rb1 cell line (W) and in frozen retinoblastoma samples (8–10). Antiactin has been used as loading control. (b, c) Immunohistochemical analysis of pRb2/p130 in normal retina (b) and in a representative retinoblastoma sample (c). (d–f) Laser capture microdissection of two selected areas (e, f) of paraffin-fixed tumor sample (d). (g) Exon 1 homozygous missense mutations at codons 178 and 259 detected in a representative DNA sample obtained by laser capture microdissection. The sequences were matched with the wild-type sequences (Gene Bank Accession Numbers X74594 and U53220)
Role of epigenetic events on Rb2 / p130 gene expression
CpG islands, which are potential methylation sites, are often found near the promoter of widely expressed genes and typically extend into the first exon. CpG islands can also occur downstream from transcription start sites and are unmethylated in normal cells, although they seem to be preferential targets for de novo methylation in human cancer.
Therefore, we processed all samples by methylation specific-PCR (MSP) assay focusing our analysis on RB2/p130 regions rich in CpG dinucleotide: the promoter region immediately 5′-flaking to the transcription start site (ATG), Exon 1 and Intron 1 (Figure 2a and Table 2b). In Figure 2b, we report the results obtained in three samples chosen as representative of pRb2/p130 expression level pattern (Table 1: samples 2, 3 and 8). When pRb2/p130 expression level is normal (+++), all the three examined regions were unmethylated (U1, U2 and U3). Loss of expression (−) corresponded to methylation of the regions 1, 2 and 3 (M1, M2 and M3), while downregulation (+) was correlated to methylation of region 3 alone (Table 1). These results suggest that Exon 1 mutation pattern could establish the susceptibility to gene methylation which, in turn, would determine the protein expression level.
Rb2/p130 methylation status with respect to pRb2/p130 expression levels. (a) Schematic representation of CpG sites on the Rb2/p130 promoter, Exon 1 and Intron 1. The three examined regions are indicated. Region 1 spans from −95 bp to +177 bp, region 2 from +167 bp to +302 bp and region 3 from +287 bp to +411 bp. (b) MSP of Rb2/p130 CpG rich regions of three representative samples showing different pRb2/p130 expression levels. MSP amplification of modified DNA was performed with specific primers for methylated (M1, M2 and M3) and unmethylated (U1, U2 and U3) DNAs. Retinoblastoma unmodified (C1, positive control) and modified (C2, negative control) DNAs amplified with wild-type primers. The sample with normal pRb2/p130 expression (+++) shows the three regions as being unmethylated (U1, U2 and U3); the sample with weak expression (+) shows only region 3 (M3) as being methylated and the sample with negative expression (−) shows all three regions as being methylated (M1, M2 and M3)
To verify whether removal of the transcriptional block due to methylation changes the expression level of endogenous pRb2/p130 and restores its growth suppressive function on cell proliferation, we treated Weri-Rb1 cell line with the DNA methyltransferase inhibitor 5-Aza-2-deoxycytidine (5-Aza-dC). Actually, data obtained in Weri-RB1 cell line indicate that, in this model, percentage of methylation and therefore of Rb2/p130 expression level are not so much univocally correlated with mutational status as we have found in primary tumors. However, we felt it opportune to investigate the effects of demethylating agents in the sole, even if imperfect, experimental model available.
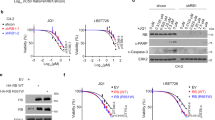
The effect of the demethylating agent on Weri-Rb1 cell proliferation as a function of treatment duration is shown in Figure 3a. As evident, a significant difference in proliferation rate was detectable after 96 h of treatment when the number of cells was markedly reduced. FACS analysis (data not shown) revealed that this reduction in total cell number corresponds to an increase in the amount of apoptotic cells (20% of treated cells vs 6% of control). Moreover, preponderance of G1 arrested cells was already detected after 24 h (40% of treated cells vs 30% of control). Western blot analysis showed that the effects of the de-methylating treatment on cell growth were concomitant with increased expression of endogenous pRb2/p130 with respect to the control obtained after 96 h cell culture (Figure 3b). Recently, we reported that pRb2/p130 is involved in p53-independent apoptotic response via a p73 pathway (La Sala et al., 2003). Therefore, we evaluated whether the apoptotic response evidenced by FACS analysis is mediated by this same pathway by analysing the expression level of E2F1 and p73 as a function of the treatment period. Moreover, we assessed also the expression level of E2F4, necessary for pRb2/p130 antiproliferative function, and p53, which was shown to be downregulated in this cell line but whose expression increased, inducing an apoptotic response, after γ-irradiation treatment (Kondo et al., 1997). After 48 h 5-Aza-2-dC treatment, pRb2/p130, p73, E2F1 and p53 upregulation was evident and reached a maximum after 96 h while E2F4 expression level resulted unchanged (Figure 3b). These results indicate that both p53-dependent and p73-dependent apoptotic pathways could synergistically participate in leading the cells to programmed cell death.
Effect of 5-Aza-dC treatment on Weri-Rb1 cell line. (a) Proliferation analysis of Weri-Rb1 cell line treated with 2.5 μ M 5-Aza-2-dC at different times (24, 48 and 96 h). The proliferation index was calculated as a percentage of the signal of sample relative to the signal of untreated cells at 0 h. (b) Western blot analysis of pRb2/p130, E2F1, E2F4, p73 and p53 using whole cell lysates from Weri-Rb1 cell line treated with 2.5 μ M 5-Aza-2-dC at different times (24, 36, 48, 72 and 96 h). Whole cell lysates from untreated Weri-Rb1 cells harvested at 96 h were used as control (C). Equal concentration of 100 μg of total proteins has loaded in each well (Actin)
Discussion
A thorough comprehension of the multiplicity of molecular mechanisms leading to cancer is becoming more and more necessary for both prevention and therapy. Moreover, it now seems evident that for each tumor type we can distinguish various subclasses reflecting different molecular backgrounds. At the same time, every tumor can be ascribed to a restricted number of events among which loss of control on cell proliferation and disruption of the apoptotic response play a crucial role (Hanahan and Weinberg, 2000). Recently, the concept that epigenetic, as well as genetic, events might be central to the evolution of human cancer is re-emerging. Cancer often exhibits an aberrant methylation of gene promoter regions, which is associated with loss of gene function (Sugimura and Ushijima, 2000). Overall, these considerations lead to the conclusion that an accurate characterization of the molecular events typical of each tumor is fundamental. Retinoblastoma is a rare malignant tumor of the developing retina occurring in familial (10% of cases) or sporadic form (90% of cases). Children with mutations in the RB1/p105 gene are specifically susceptible to retinoblastoma even if it has been recently reported that loss of RB1/p105 from a developing retinal cell is insufficient for malignancy (Gallie et al., 1999). We previously reported that 14 out of 40 sporadic retinoblastomas showed loss of pRb2/p130 expression, while the remaining cases showed a reduction in pRb2/p130 expression (median 40%). Interestingly, loss of expression correlated with a lesser degree of cellular differentiation and low apoptotic index (Bellan et al., 2002). pRb2/p130, which is as well expressed in normal retina, shares with pRb1/p105 both antiproliferative and proapoptotic functions even if they act through distinct pathways.
The most direct evidence of pRb/p105 control on p53 proapoptotic activity is its ability to prevent p53 degradation by forming a trimeric complex with MDM2 (Hsieh et al., 1999). Interestingly, pRb/p105 acetylation is needed for pRb/p105-MDM2 binding (Chan et al., 2001).
Moreover, pRb/p105 deregulation determines unrestrained E2F1 expression, which may cause p53-dependent and -independent apoptosis (Hsieh et al., 1997; Johnson and Schneider-Broussard, 1998). Namely, E2F1 has been shown to induce the accumulation of p53 (Hiebert et al., 1995; Kowalik et al., 1998), the transcription of p73 in cells lacking p53 (Irwin et al., 2000; Stiewe and Putzer, 2000) and the transcription of Apaf-1, a key element of apoptosome (Moroni et al., 2001).
In addition, it has been reported that acetylation, dephosphorylation and consequent ubiquitin-mediated degradation of pRb/p105 could be early events in programmed cell death, possibly related to cell commitment to apoptosis (Cinti et al., 2003).
On the other hand, pRb2/p130-E2F4 complex negatively modulates the E2F1 promoter (Johnson, 1995), suggesting that pRb2/p130 indirectly modulates p73 expression by controlling E2F1 transcription. Moreover, it has been recently reported that pRb2/p130 is also able to control p73 transcription directly by forming distinct multimolecular complexes on p73 promoter in cells, which lack both pRb1/p105 and p53 (Cinti et al., 2003). These findings suggest that both pRb1/p105 and pRb2/p130 can share proapoptotic function by activating distinct pathways and the involvement of one or the other depends on their functional status. Actually, tumors concomitantly lacking pRb/p105 and p53, such as osteosarcoma and retinoblastoma, are still able to undergo apoptosis, thus suggesting a possible role in this process of some other pRB protein, most probably pRb2/p130.
Therefore, the assessment of RB2/p130 mutational status in an RB1/p105-deleted scenario could be relevant to better understand genesis and progression of retinal cancer.
We have found two Exon 1 mutations of RB2/p130 gene present in all the examined sporadic retinoblastomas. It seems that the effect of each mutation on the protein expression level depends on loss of heterozygosity and the two of them are needed to abolish pRb2/p130 expression. Interestingly, one of the familial retinoblastomas shows both mutations but heterozygosity is sufficient to prevent any effect. This result, when confirmed in a larger number of cases, would offer a possibility to assume the gene mutational status from immunohistochemistry.
Exon 1 is a CpG-enriched region and, therefore, it is susceptible to being improperly methylated. On the other hand, it has been reported that also RB1/p105 gene was found hypermethylated in retinoblastoma (Stirzaker et al., 1997). Screening of methylation status in RB2/p130 gene promoter shows that a 178 (TCT → CCT) homozygous mutation in Exon 1 correlates with hypermethylation at one site determining a reduction in protein expression level. Actually, this mutation alters susceptibility to methylation of a region (M3) bridging Exon 1 and Intron 1, thus allowing transcription even if it is impaired. When a 259 (CCC → GCC) homozygous mutation adds to the other one, promoter as well Exon 1 and Intron 1 regions become aberrantly methylated and transcription is abolished.
This result indicates that pRb2/p130 could participate in human sporadic retinoblastoma onset. In the mouse, RB1/p105 loss is insufficient to cause retinoblastoma (Maandang et al., 1994), while chimeras composed of cells lacking both RB1/p105 and p107 develop retinal cancer (Robanus-Maandang et al., 1998). The effects of loss of both RB1/p105 and p107 have been recently confirmed and extended in p107−/− mice after conditional inactivation of RB1/p105 in the retina (Chen et al., 2004). In particular, it was shown that a subclass of retinal inner nuclear layer precursors survive RB1/p105 or RB1/p105 and p107 loss, divide ectopically and terminally differentiate in amacrine cells by postnatal day 30 (P30). Retinoblastomas displaying similar histology, and likely histogenesis, to human retinal cancer develop also when RB1/p105 loss in the retina is induced on a pRb2/p130-deficient background (MacPherson et al., 2004).
Overall, these authors' results enlighten the tight cooperation among the three retinoblastoma family proteins during murine retinal development. In the absence of pRb1/p105, pRb2/p130 and p107 can compensate its functions by modulating their growth suppressive activity more than their expression level. Namely, on an RB1/p105-deficient background, and even more when also pRb2/p130 is absent, p107 is present in its active underphosphorylated form and this is most probably due to cyclin D1 downregulation. On the other hand, it seems that pRb1/p105 and pRb2/p130 could play some cell-specific role in different retinal neuron precursors since pRb1/p105 loss leads to profound effects on retinal ganglion cells, photoreceptors and bipolar cells while horizontal cells, Mueller glia and amacrine cells survive. The role of retinoblastoma family proteins in human retinal development is still unknown, but our data on sporadic retinoblastoma suggest that, even if pRb2/p130 and/or p107 are not able to compensate for RB1/p105 deletion in human retina, impairment of one of them may most probably contribute to cancer onset.
Weri-Rb1 cell line imperfectly models what we observed in primary retinoblastoma. In particular, not withstanding the fact that both Exon1 mutations are homozygous, RB2/p130 gene is only partially methylated. However, cell lines are a unique tool for investigating the effects of demethylating agents and we chose Weri-Rb1 cell line because it presents complete biallelic deletion of RB1/p105, while Y79 cell line contains a partial deletion at one allele and an uncharacterized mutation in the other (Kondo et al., 1997). Therefore, we analysed the effects in time of 5-Aza-dC on both cell proliferation and pRb2/p130 expression level in Weri-Rb1 cell line. Moreover, we also screened changes in the expression level of proteins involved in both pRb2/p130 prosurvival (E2F4) and proapoptotic pathways (E2F1 and p73) and of the proapoptotic protein p53 whose expression is altered in the retinoblastoma without any mutation (Kondo et al., 1997; Gallie et al., 1999). The number of viable cells 96 h after a single 5-Aza-dC treatment was about halved, and FACS analysis showed that this reduction could be ascribed to both G1 arrest and apoptotic response. At the same time pRb2/p130 expression level was maximum. Recently, we have shown that pRb2/p130 is able to induce p53-independent apoptosis by controlling the transcription of p73 through the binding of activating pRb2/p130-E2F1-p300 or repressive pRb2/p130-E2F4-HDAC1 multimolecular complexes on p73 promoter in osteosarcoma (La Sala et al., 2003). Here we show that an increase in the expression of pRb2/p130 is concomitant with upregulation of E2F1 and p73 but not of E2F4, suggesting that activation of the proapoptotic pathway could play a preponderant role in determining growth cell arrest. Interestingly, a model of retinoblastoma genesis that takes into account the centrality of programmed cell death in normal retinal development has been recently proposed (Gallie et al., 1999). The authors suggest that the additional mutations that are present in all retinoblastomas may interrupt signals that normally would induce apoptosis when RB1/p105 is absent. Actually, our previous data (La Sala et al., 2003) were obtained in the Hos osteosarcoma cell line, which presents RB1/p105 biallelic deletion, and osteosarcoma is the most common neoplasm secondary to retinoblastoma (Eng et al., 1993). Therefore, RB2/p130 inactivation could contribute to retinoblastoma genesis, depriving the cell of a further tool to control proliferation and to induce the elimination of inappropriate cells by a p73-dependent proapoptotic pathway. A few evidences support this hypothesis. pRb2/p130 upregulation during development and maturation of nervous system, including the retina, has been reported (Kastner et al., 1998; Kusek et al., 2001). Nothing is known about p73 role in modeling the precise retinal architecture, but it has been reported that, if p53 responds to immediate genotoxic stress, its homologue p73 would participate in the cell homeostasis of neuronal tissue along its development and differentiation (Douc-Rasy et al., 2004). Interestingly, MacPherson et al. (2004) reported that apoptosis in mouse RB/p105−/− retinas is mainly, even if not exclusively, p53-independent and a direct consequence of RB/p105 deletion.
Our data suggest that pRb2/p130 loss of expression could interfere with the activation of the p53-independent/p73-dependent proapoptotic pathway, thus determining death resistance in some RB/p105-deficient retinal neurons. Actually, in mice mutant for RB/p105 and RB2/p130, retinoblastoma may be explained by the survival of some amacrine cells. On the other hand, treatment of Weri-Rb1 with a 5-Aza-dC also induced upregulation of p53, which has been shown to activate p53-dependent apoptotic pathway in this cell line (Kondo et al., 1997). No p53 mutation has been detected in human retinoblastoma (Gallie et al., 1999) even if it is not possible to rule out that its expression could be hampered by epigenetic mechanisms as it seems the case in Weri-Rb1 cell line.
Unfortunately, although treatment strategies for cancer have gradually evolved throughout the past decades, enucleation and radiotherapy are the most common retinoblastoma treatments and salvage of useful vision is possible only in limited cases (Lai et al., 2003). Recently, a proposed international clinical trial has been evaluating systemic chemotherapy (etoposide, carboplatin and vincristine) to achieve local tumor volume reduction and prevent other tumor formation. Since CpG island methylation is not commonly found in normal cells, cytosine methylation provides a highly specific target for pharmacological therapy. Inhibition of cytosine methylation by pharmacological or biological strategies has been shown to prevent or reverse the onset of neoplasia in various animal models (Bender et al., 1998; Kalibic, 2003). Ongoing clinical trials are evaluating the safety and efficacy of agents affecting epigenetic changes in cancer patients (Brown and Strathdee, 2002). Our data provide new information about the genetic basis and the contribution of aberrant hypermethylation in sporadic retinoblastoma, the most frequent form of child eye cancer, suggesting that treatment with demethylating agents could help for a successful therapy.
Materials and methods
Case selection and processing of tissue for histological evaluation
The paraffin wax blocks of a total of 10 pretreatment surgical specimens of ocular retinoblastomas and three normal retina samples, obtained from a patient who underwent enucleation for painful absolute glaucoma, were collected at the Deparment of Human Pathology and Oncology, University of Siena, Italy. Tissues were cut and fixed in a buffered 4% aqueous formaldehyde solution, pH 7.4. For conventional histology, 4-μm-thick sections were obtained from representative paraffin wax blocks and stained with hemalum and eosin, Giemsa, periodic acid-Schiff (PAS), Gomori's silver impregnation and Feulgen.
Immunohistochemistry
Consecutive sections of retinoblastoma tissue cut at 3-μm thickness were subjected to immunostaining. Tissue samples of normal retina from three unrelated patients were used as the control. We used the EnVision™ +/HRP method (Dako, Milan, Italy) to visualize immunohistochemical reaction products. Antigen retrieval was achieved by the treatment of deparaffinized sections with microwaves in 1 mM EDTA, pH 8.0, for 5 min, followed by cooling at room temperature prior to incubation with the antibodies. The monoclonal antibody anti-pRb2/p130 was obtained from Transduction Laboratories (Lexington, KY, USA), and was used at a dilution in TBS of 1 : 100. We replaced the primary antibodies with normal mouse serum to obtain negative controls. Normal human tonsils served as positive controls.
Laser capture microdissection and DNA extraction
Three normal human retina and seven retinoblastoma archived paraffin-fixed tissues were identified on hematoxylin and eosin (H&E)-stained sections and isolated by laser capture microdissection (Arcturus PixCell II™ MWG-BIOTECH, Florence, Italy). The selected samples adhered to a Capsure™ transfer film. The Capsure™ transfer film carrier was placed directly onto a standard microcentrifuge tube containing digestion buffer (50 μl buffer containing 0.04% Proteinase K, 10 mM Tris-HCL (pH 8.0), 1 mM EDTA and 1% Tween-20). The tube was preheated upright in a 37°C oven for 5 min and then placed upside down so that the digestion buffer contacted the tissue on the cap. The samples were incubated overnight at 37°C, centrifuged for 5 min and the cap was removed. The samples were heated to 95°C for 8 min to inactivate the Proteinase K and used directly as template for PCR reactions. Genomic DNAs were extracted from three frozen retinoblastoma samples, Weri-Rb1 cell line and 15 health donors blood samples according to the standard manufacturer's instructions.
pRb2 / p130 mutational analysis and methylation-specific PCR (MSP) assay
PCR of genomic DNA extracted from microdissected primary tumors and Weri-Rb1 cell line was performed for mutational analysis of Rb2/p130. All 22 exons (Table 2a) were amplified at an annealing temperature of 55°C and sequenced. Moreover, we examined the status of methylation of the 10 retinoblastoma specimens and Weri-Rb1 cell line in the density of CpG immediately 5′ to the transcription started site (ATG) and inside Exon 1 and Intron 1. These regions were identified by using the ‘CpG Ware ™ primer design software’ (Intergen, Purchase, NY, USA). The assay is based on the DNA sequence differences between methylated and unmethylated DNA after bisulfite modification by DNA modification CpGenome Kit (Intergen, Purchase, NY, USA). The bisulfite reactions were performed according to the manufacturer's instructions. Subsequent PCR with primers specifically designed for discriminating between methylated (Tm=66°C), unmethylated (Tm=61°C) and wild-type DNA (Tm=68°C) was performed (Table 2b). PCR products were analysed on 2.5% agarose gel.
Culture cell line and 5-Aza-2-dc DNA methyltransferase inhibitor treatment
Human retinoblastoma cell line (Weri-Rb1) obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) was cultured in RPMI 1640 supplemented with 10% FCS at spin ratio of 1 : 2 once a week. A volume of 2.5 μ M of 5-Aza-2-dC, a DNA methyltransferase inhibitor, (Sigma-Aldrich, St Louis, MO, USA) was added to medium culture of treated cells.
Cell viability (MTT) and FACS analysis
Quantitative cell viability was measured by colorimetric assay using a cell proliferation kit (MTT) (Roche Molecular Biochemicals, Mannheim, Germany). A total of 5000 cells/well Weri-Rb1 and 5-Aza-2-dC-treated Weri-Rb1 were grown in microtiter plates (96-well) in a final volume of 100 μl culture medium. The incubation period of cell culture was 24, 48 and 96 h in the presence or absence of the 2.5 μ M DNA methyltransferase inhibitor. After incubation period, 10 μl MTT labelling reagent was added to each well (final concentration 0.5 μg/ml). MTT is cleaved by growing cells to form purple formazan crystals and allows quantification of cell viability by spectrophotometric analysis (ELISA) at 550 nm. Cell viability was expressed as the percentage of the absorbance of drug-treated and untreated cells relative to that of the untreated cells at 0 h. FACS analysis was carried out on cells treated with 5-Aza-2-dC and compared to those untreated (control) after 24, 36, 48, 72 and 96 h of culture.
Western blotting
Western blotting analysis of pRb/p105, pRb2/p130 was performed on three fresh samples of primary tumors (samples 8–10), which resulted in downregulation of pRb2/p130 by immunohistochemistry. Weri-Rb1 cell line and one normal retina sample, obtained from a patient who underwent enucleation for painful absolute glaucoma, served as an experimental control. Fresh tissues were immediately frozen in liquid nitrogen. Furthermore, Weri-Rb1 cells treated and untreated with 5-Aza-2-dC at various times were processed for Western blot analysis. Whole tissue lysates were prepared by resuspending homogenized tissues and pelleted cells in lysis buffer (50 mM Tris/HCl, 5 mM EDTA, 250 mM NaCl, 50 mM NaF, 0.1% Triton X-100, 0.1 mM Na3VO4 plus fresh inhibitors). Equal amounts of 100 μg of total extracts were loaded and resolved on a 7 or 10% SDS–PAGE. The gels were then transferred onto a nitrocellulose filter and checked by using 0.1% Ponceau red. The polyclonal anti-pRb/p105 (C15) (Santa Cruz, Santa Cruz, CA, USA) and monoclonal anti-pRb2/p130 (Transduction Laboratories, Lexington, KY, USA) were used at a dilution of 1 : 300 and 1 : 500, respectively. The monoclonal anti-p53 (Ab6) (Calbiochem, Cambridge, MA, USA) was used at a dilution of 1 : 1000, while the monoclonal anti-E2F1 (KH95), polyclonal anti-E2F4 (C20) and polyclonal anti-p73 (H79) (Santa Cruz, Santa Cruz, CA, USA) were used at dilution of 1:200. The anti-actin antibody (Santa Cruz, Santa Cruz, CA, USA) was used as loading control following the manufacturer's instructions.
References
Baldi A, Boccia V, Claudio PP, De Luca A and Giordano A . (1996). Proc. Natl. Acad. Sci. USA, 93, 4629–4632.
Bellan C, De Falco G, Tosi GM, Lazzi S, Ferrari F, Morbini G, Bartolomei S, Toti P, Mangiavacchi P, Cevenini G, Trimarchi C, Cinti C, Giordano A, Leoncini L, Tosi P and Cottier H . (2002). Invest. Ophthalmol. Visual Sci., 43, 3602–3608.
Bender CM, Pao MM and Jones PA . (1998). Cancer Res., 58, 95–101.
Brown R and Strathdee G . (2002). Trends Mol. Med., 8, S34–S48.
Chan HM, Krstic-Demonacos M, Smith L, Demonacos C and La Thangue NB . (2001). Nat. Cell Biol., 3, 667–674.
Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M and Bremner R . (2004). Cancer Cell, 5, 539–551.
Choy KW, Pang CP, Fan DS, Lee TC, Wang JH, Abramson DH, Lo KW, To KF, Yu CB, Beaverson KL, Cheung KF and Lam DS . (2004). Invest. Ophthalmol. Visual Sci., 45, 3404–3409.
Cinti C and Giordano A . (2000). Emerging Ther Targets, 6, 765–783.
Cinti C, Trimarchi C and Giordano A . (2003). Regulation of G1 Phase Progression. Boonstra J (ed). Springer-Verlag: Berlin/Heidelberg/New York, pp 200–235.
DiCiommo D, Gallie BD and Bremner R . (2000). Semin. Cancer Biol., 10, 255–269.
Douc-Rasy S, Goldschneider D, Million K and Benard J . (2004). Med. Sci. (Paris), 20, 317–324.
Eng C, Li FP, Abramson DH, Ellsworth RM, Wong FL, Goldman MB, Seddon J, Tarbell N and Boice Jr JD . (1993). J. Natl. Cancer Inst., 85, 1121–1128.
Gallie BL, Campbell C, Devlin H, Duckett A and Squire JA . (1999). Cancer Res., 59, 1731–1735.
Hanahan D and Weinberg RA . (2000). Cell, 100, 57–70.
Hiebert SW, Packham G, Strom DK, Haffner R, Oren M, Zambetti G and Cleveland JL . (1995). Mol. Cell. Biol., 15, 6864–6874.
Hsieh JK, Fredersdorf S, Kauzarides T, Martin K and Lu X . (1997). Genes Dev., 11, 1840–1852.
Hsieh JK, Chan FS, O'Connor DJ, Mittnacht S, Zhong S and Lu X . (1999). Mol. Cell., 3, 181–193.
Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W, Flores ER, Tsai KY, Jacks T, Vousden KH and Kaelin Jr WG . (2000). Nature, 407, 645–648.
Issa JP . (1999). Crit. Rev. Oncol. Hematol, 32, 31–43.
Johnson DG . (1995). Oncogene, 11, 1685–1692.
Johnson DG and Schneider-Broussard R . (1998). Front. Biosci., 3, d447–d448.
Kalibic T . (2003). Ann. NY Acad. Sci., 983, 278–285.
Kastner A, Espanel X and Brun G . (1998). Cell Growth Differ., 9, 857–867.
Knudson AG . (2002). Am. J. Med. Genet., 111, 96–102.
Knudson Jr AG, Hethcote HW and Brown BW . (1975). Proc. Natl. Acad. Sci. USA, 72, 5116–5120.
Kondo Y, Kondo S, Liu J, Haqqi T, Barnett GH and Barna BP . (1997). Exp. Cell Res., 236, 51–56.
Kowalik TF, DeGregori J, Leone G, Jakoi L and Nevins JR . (1998). Cell Growth Differ., 9, 113–118.
Kusek JC, Greene RM and Pisano MM . (2001). Brain Res. Bull., 54, 187–198.
La Sala D, Macaluso M, Trimarchi C, Giordano A and Cinti C . (2003). Oncogene, 22, 3518–3529.
Lai H, Ma F and Lai S . (2003). J. Cell. Biochem., 88, 121–127.
Maandang EC, van der Valk M, Vlaar M, Feltkamp C, O'Brien J, van Roon M, van der Lugt N, Berns A and te Riele H . (1994). EMBO J., 13, 4260–4268.
MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME and Jacks T . (2004). Genes Dev., 18, 1681–1694.
Mayol X, Grana X, Baldi A, Sang N, Hu Q and Giordano A . (1993). Oncogene, 8, 2561–2566.
Moroni MC, Hickman ES, Denchi LE, Caprara G, Colli E, Cecconi F, Muller H and Helin K . (2001). Nat. Cell. Biol., 3, 552–558.
Robanus-Maandang E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, Berns A and te Riele H . (1998). Genes Dev., 12, 1599–1609.
Stiewe T and Putzer BM . (2000). Nat. Genet., 26, 464–469.
Stirzaker C, Millar DS, Paul CL, Warnecke PM, Harrison J, Vincent PC, Frommer M and Clark SJ . (1997). Cancer Res., 57, 2229–2237.
Sugimura T and Ushijima T . (2000). Mutat. Res., 462, 235–246.
Zheng L and Lee WH . (2002). Adv. Cancer Res., 85, 13–50.
Acknowledgements
This work was supported by AIRC, Fondazione MPS, MURST-LAG-CO3 and CNR Italian grants; Sbarro Health Research Organization (www.shro.org) and NIH USA-grants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tosi, G., Trimarchi, C., Macaluso, M. et al. Genetic and epigenetic alterations of RB2/p130 tumor suppressor gene in human sporadic retinoblastoma: implications for pathogenesis and therapeutic approach. Oncogene 24, 5827–5836 (2005). https://doi.org/10.1038/sj.onc.1208630
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1208630
Keywords
This article is cited by
-
Epigenetic regulation of human retinoblastoma
Tumor Biology (2016)
-
Epigenetic and Copy Number Variation Analysis in Retinoblastoma by MS-MLPA
Pathology & Oncology Research (2012)
-
Rb2/p130 is the dominating pocket protein in the p53–p21 DNA damage response pathway leading to senescence
Oncogene (2009)
-
A small molecule based on the pRb2/p130 spacer domain leads to inhibition of cdk2 activity, cell cycle arrest and tumor growth reduction in vivo
Oncogene (2007)
-
pRb2/p130: a new candidate for retinoblastoma tumor formation
Oncogene (2006)