Abstract
Aberrant promoter methylation of tumor suppressor genes has not been fully investigated in pediatric tumors. Therefore, we examined the methylation status of nine genes (p16INK4A, MGMT, GSTP1, RASSF1A, APC, DAPK, RARβ, CDH1 and CDH13) in 175 primary pediatric tumors and 23 tumor cell lines using methylation-specific PCR. We studied the major forms of pediatric tumors – Wilms' tumor, neuroblastoma, hepatoblastoma, medulloblastoma, rhabdomyosarcoma, osteosarcoma, Ewing's sarcoma, retinoblastoma and acute leukemia. The most frequently methylated gene in both primary tumors and cell lines was RASSF1A (40, 86%, respectively). However, the rates of RASSF1A methylation in individual tumor types varied from 0 to 88%. RASSF1A methylation was tumor specific and was absent in adjacent non-malignant tissues. Methylation of the other genes was relatively rare in tumors and non-malignant tissues (less than 5%). Neuroblastoma patients with methylation of RASSF1A were significantly older than patients without methylation (P=0.008). There was no relationship between methylation status and other clinico-pathologic parameters. We treated six cell lines lacking RASSF1A mRNA with 5-aza-2′deoxycytidine to examine the relationship between methylation and transcriptional silencing. In five of six cell lines, restoration of RASSF1A mRNA was confirmed by RT–PCR. Our findings indicate that aberrant promoter methylation of RASSF1A may contribute to the pathogenesis of many different forms of pediatric tumors.
Similar content being viewed by others
Main
Pediatric solid tumors develop after relatively short latent periods and, in contrast to the common adult epithelial tumors (Loeb, 2001), contain relatively few mutations (Davidoff and Hill, 2001). Aberrant methylation of normally unmethylated CpG islands located in the promoter regions is associated with transcriptional inactivation of defined tumor suppressor genes (TSGs) in human cancers (Baylin et al., 1998). Thus, aberrant promoter methylation may contribute to the pathogenesis and progression of malignant tumors. The methylation status of adult tumors has been studied extensively and each tumor type appears to have a distinct methylation profile (Esteller et al., 2001). By contrast, there is less information about methylation in childhood malignancies. While reports exist of the methylation of individual genes in Wilms' tumor (Mares et al., 2001), neuroblastoma (Astuti et al., 2001; Takita et al., 2000; Teitz et al., 2000), rhabdomyosarcoma (Chen et al., 1998), medulloblastoma (Fruhwald et al., 2001a,b), retinoblastoma (Ohtani-Fujita et al., 1997) and acute lymphoblastic leukemia (Nakamura et al., 1999; Wong et al., 2000), a methylation profile of the spectrum of pediatric tumors using a panel of genes has not been published.
We studied the methylation profiles of 175 childhood tumors, including most of the major solid types. Tumors were predominantly obtained from Children's Hospital Medical Center, (Dallas, TX, USA) after obtaining Institutional Review Board approval and informed consent. Some of the hepatoblastomas were obtained from the Pediatric Oncology Group Hepatoblastoma Tumor Bank. The retinoblastomas were from the University of Siena (Siena, Italy). Twenty-three pediatric tumor cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA). We examined the status of nine genes frequently showing promoter region methylation in adult tumors. The genes included RASSF1A (Ras association domain family 1, isoform A), a newly described 3p21.3 TSG, p16INK4A, MGMT (O6-methylguanine DNA methyltransferase), GSTP1 (glutathione S-transferase P1), APC (adenomatous polyposis coli), DAPK (death-associated protein kinase), RARβ (retinoic acid receptor-β), CDH1 (E-cadherin) and CDH13 (H-cadherin) and they have been investigated extensively in lung, breast, colon and prostate and other adult cancers (Burbee et al., 2001; Esteller et al., 1998, 1999a,b; Graff et al., 1997; Herman et al., 1996; Maruyama et al., 2001, 2002; Sato et al., 1998; Toyooka et al., 2001a; Tsuchiya et al., 2000; Virmani et al., 2000).
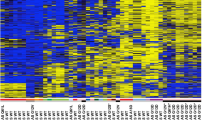
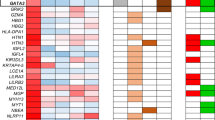
Aberrant promoter methylation was examined using methylation-specific PCR (MSP) (Herman et al., 1996). RASSF1A was methylated in 70 of 175 (40%) primary tumors and in 20 of 23 (86%) of the cell lines (Figure 1, Table 1, Table 2). However, methylation frequencies of other genes were relatively low in tumors. In general, methylation frequencies of these genes were also low in the cell lines, although CDH1 was methylated in 43% of cell lines (compared to 4% of tumors). The frequencies of methylation of RASSF1A varied with the tumor type, being highest in medulloblastoma (88%), rhabdomyosarcoma (61%), retinoblastoma (59%), neuroblastoma (52%) and Wilms' tumor (42%) samples. RASSF1A methylation rates were much lower in hepatoblastoma (19%) and acute leukemia (15%), and absent in osteosarcoma and Ewing's sarcoma. Rhabdomyosarcoma and acute leukemia are classified into three and two major subtypes, respectively. In rhabdomyosarcoma samples, the RASSF1A methylation rates of alveolar, embryonal and anaplastic type were four of six, five of seven and none of two, respectively. In leukemias, 17% of acute lymphoblastic leukemias were methylated, but methylation was absent in acute myelogenous leukemias. In contrast to the highly malignant neuroblastomas, RASSF1A methylation was absent in ganglioneuromas (n=6), which are benign tumors consisting of ganglion and Schwann cells but lacking progenitor neuroblast cells. Methylation was absent in corresponding non-malignant tissues except for p16INK4A methylation in one histologically normal kidney.
Methylation-specific PCR of RASSF1A in primary pediatric tumors. The methylated forms in representative samples of each tumor type are illustrated. The MSP product size of methylated RASSF1A was 169 bp. Primary pediatric tumors (n=175) and 23 cell lines (Table 2) were examined by MSP to determine methylation status of nine genes. Rhabdomyosarcoma samples included alveolar (n=6), embryonal (n=7) and anaplastic type (n=2). Eighteen acute lymphoblastic (ALL) and two acute myelogenous leukemia (AML) samples were included in acute leukemia. Non-malignant tissues included corresponding normal kidney (n=11) and liver (n=1) tissues, and normal adult lymphocytes (n=2). Genomic DNA was isolated from frozen tissues and cell lines by sodium dodecyl sulfite (SDS) and proteinase K digestion, phenol/chloroform extraction and ethanol precipitation. DNA was treated with sodium bisulfite as described previously (Herman et al., 1996). Briefly, 1 μg of DNA was denatured by 0.2 M NaOH for 10 min at 37°C. Thirty μl of 10 mM hydroquinone (Sigma Chemical Co., St. Louis, MO, USA) and 520 μl of 3M sodium bisulfite (Sigma Chemical Co.) at pH 5.0 were added and mixed, and then the solution was incubated at 50°C for 16 h. Treated DNA was purified using the Wizard DNA clean-up system (Promega Co., Madison, WI, USA). The DNA was precipitated by ethanol and used immediately or stored at −70°C until used. Two μl of bisulfite-modified DNA was amplified by PCR using primers that were specific for methylated or unmethylated sequences of each genes as described previously (Burbee et al., 2001; Esteller et al., 1998, 1999a,b; Graff et al., 1997; Herman et al., 1996; Sato et al., 1998; Toyooka et al., 2001a; Tsuchiya et al., 2000; Virmani et al., 2000). The unmethylated form of p16INK4A was examined as a control for DNA integrity and was present in all samples. PCR amplified products were electrophoresed on 2% agarose gel and visualized under ultraviolet illumination. Letters above indicate WT, Wilms' tumor; NB, neuroblastoma; HB, hepatoblastoma; RMS, rhabdomyosarcoma; MB, medulloblastoma; OS, osteosarcoma; EWS, Ewing's sarcoma; RB, retinoblastoma; AL, acute leukemia; NT, non-malignant tissues; M, marker; P, positive control; N, negative control (water blank)
We also examined the relationship between methylation of RASSF1A and clinico-pathologic parameters including sex, age, stage, existence of metastasis and outcome. Statistical analyses for differences between groups were performed using χ2, Fisher's exact test and Mann–Whitney U-test. The Kaplan–Meier log-rank test was performed for analysis of overall survival. Neuroblastoma patients with methylation of RASSF1A were significantly older than patients whose tumors lacked methylation (2.25±1.65 years old, vs 0.94±1.10 years old, respectively; uncorrected P value=0.008, Mann–Whitney U-test). Wilms' tumor, rhabdomyosarcoma, medulloblastoma and retinoblastoma patients with methylation of RASSF1A also tended to be older compared to patients without methylation, but the differences were not significant. Larger studies will be required to conclusively demonstrate that methylation of RASSF1A in pediatric tumors is related to patient age. There was no relationship between methylation status and other clinico-pathologic parameters including survival.
RASSF1A, located on chromosome 3p21.3, has been demonstrated to function as a TSG in lung cancer (Burbee et al., 2001; Dammann et al., 2000) and is inactivated in many other tumors including breast, renal, bladder and ovarian carcinomas and malignant mesothelioma (Burbee et al., 2001; Dreijerink et al., 2001; Morrissey et al., 2001; Toyooka et al., 2001b; Yoon et al., 2001). The gene encodes two major transcripts which are produced by alternative promoter selection and alternative mRNA splicing: RASSF1A (340 amino acids, encoding a 39-kd peptide) containing a predicted N-terminal diacylglycerol-binding (DAG) domain and a C-terminal predicted RAS associated domain; and RASSF1C (270 amino acids, encoding a 32-kd peptide) with a different N-terminus lacking the DAG domain but exhibiting a similar C-terminus containing the RAS association domain (Burbee et al., 2001; Dammann et al., 2000). Three transcripts are derived from two known promoters, and promoter 1A controls expression of transcripts 1A and 1F, while promoter 1C controls expression of transcript 1C.
Because of the high frequency of RASSF1A gene methylation in childhood tumors, we studied its expression in methylation positive and negative pediatric tumor cell lines. We selected six cell lines (two neuroblastoma, two retinoblastoma, one rhabdomyosarcoma and one medulloblastoma), and these cell lines had a methylated RASSF1A allele but lacked an unmethylated allele (Figure 2a). As in other cancers, all pediatric tumor cell lines tested expressed transcript 1C, while there was a selective loss of transcripts 1A and 1F associated with methylation of promoter 1A. In order to further investigate the relationship between methylation of RASSF1A and transcriptional silencing, we examined expression of RASSF1A mRNA by RT–PCR before and after treatment with 5-aza-2′ deoxycytidine (5-aza-CdR), a demethylating agent, in the six cell lines. No expression of transcript 1A was detected in all cell lines before 5-aza-CdR treatment (Figure 2b). After 5-aza-CdR treatment, restoration of transcripts 1A and 1F occurred in five of the six cell lines, but not in medulloblastoma cell line D283. The latter cell line appeared to be particularly sensitive to the toxic effects of 5-aza-CdR, and marked cytotoxic effects were noted at the time of harvesting. As in lung and breast cancers (Burbee et al., 2001; Dammann et al., 2001), RASSF1C expression was noted without any obvious change in intensity before or after 5-aza-CdR treatment. These results confirm that the transcriptional silencing of RASSF1A was caused by methylation. The absence of an unmethylated band in pediatric tumor cell lines having a methylated allele and accompanied by gene silencing suggests one of two possibilities: (a) one allele is methylated and the other allele lost by allelic loss; or (b) biallelic methylation has occurred. As we did not perform allelic loss studies in pediatric tumors, we cannot distinguish between these possibilities.
(a) Methylation status of RASSF1A in pediatric tumor cell lines. Six cell lines (SK-N-FI, SH-SY5Y, RD, D238, Y-79 and WERI-RB-1) were examined by MSP. All cell lines had a methylated allele (169 bp) but lacked an unmethylated allele (169 bp) of RASSF1A. The controls consisted of two cell lines previously examined and known to contain only methylated (m) or unmethylated (u) alleles. They were used as positive controls for the methylated and unmethylated forms respectively. The water blank lacked DNA and was used as a negative control (N) for both methylated and unmethylated forms. (b) Expression of mRNA of RASSF1A and RASSF1C before and after 5-aza-CdR treatment in pediatric tumor cell lines. Six cell lines mentioned above were grown in RPMI1640 (Life Technologies, Inc., Rockville, MD, USA) supplemented with 10% fetal bovine serum and incubated in 5% CO2 at 37°C. The cell lines were also treated with 5-aza-CdR at a concentration of 0.5–2 μg/ml for 5 days. Total RNA was extracted from cell lines using TRI Reagent (Molecular Research Center Inc., Cincinnati, OH, USA) following the manufacturer's instructions. Two μg of total RNA was reverse-transcripted by use of SUPERSCRIPTTMII First-Strand Synthesis (Gibco–BRL), and then 2 μl of cDNA was used for amplification of mRNA of RASSF1A and RASSF1C. Primer sequences were as described previously (Burbee et al., 2001). The PCR was performed using a 70–60°C touchdown method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), housekeeping gene, was used as a control for RNA integrity. Primer sequences were as described previously (Mollerup et al., 1999). PCR products were electrophoresed on 2% agarose gel and visualized under ultraviolet illumination. Letters above indicate M, marker; m, methylated allele; u, unmethylated allele; B, before treatment; A, after treatment; P, positive control; N, negative control (water blank)
Our report is the first methylation profile of all of the major forms of pediatric tumors with the exception of lymphomas. Our finding demonstrated that RASSF1A was methylated in 40% of all of the tumors tested, especially medulloblastoma, rhabdomyosarcoma, retinoblastoma, neuroblastoma and Wilms' tumors. High methylation rates were restricted to certain tumors, and the rates were low in hepatoblastoma, acute leukemia, osteosarcoma and Ewing's sarcoma. Methylation was absent in corresponding non-malignant tissues, confirming that methylation was tumor specific. Of interest, a recent publication found RASSF1A methylation in 55% of neuroblastomas (Astuti et al., 2001), compared to our rate of 52%. To our knowledge, RASSF1A methylation is the first molecular abnormality to be described that is common to a large and diverse group of pediatric tumors. Our findings indicate that aberrant promoter methylation of RASSF1A may contribute to tumorigenesis of several types of common pediatric tumors as well as many adult tumors.
Abbreviations
- TSG:
-
tumor suppressor gene
- RASSF1A :
-
Ras association domain family 1, isoform A
- MGMT :
-
O6-methylguanine DNA methyltransferase
- GSTP1 :
-
glutathionne S-transferase P1
- APC :
-
adenomatous polyposis coli
- DAPK :
-
death-associated protein kinase
- RARβ:
-
retinoic acid receptor-β
- CDH1 :
-
E-cadherin
- CDH13 :
-
H-cadherin
- MSP:
-
methylation-specific PCR
- RT–PCR:
-
reverse transcription-PCR
- 5-aza-CdR:
-
5-aza-2′deoxycytidine
References
Astuti D, Agathanggelou A, Honorio S, Dallol A, Martinsson T, Kogner P, Cummins C, Neumann HP, Voutilainen R, Dahia P, Eng C, Maher ER, Latif F . 2001 Oncogene 20: 7573–7577
Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP . 1998 Adv. Cancer Res. 72: 141–196
Burbee DG, Forgacs E, Zochbauer-Muller S, Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S, Sekido Y, Latif F, Milchgrub S, Toyooka S, Gazdar AF, Lerman MI, Zabarovsky E, White M, Minna JD . 2001 J. Natl. Cancer Inst. 93: 691–699
Chen B, Dias P, Jenkins III JJ, Savell VH, Parham DM . 1998 Am. J. Pathol. 152: 1071–1079
Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP . 2000 Nat. Genet. 25: 315–319
Dammann R, Yang G, Pfeifer GP . 2001 Cancer Res. 61: 3105–3109
Davidoff AM, Hill DA . 2001 Semin. Pediatr. Surg. 10: 106–118
Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD, Protopopov A, Li J, Kashuba V, Klein G, Zabarovsky ER . 2001 Proc. Natl. Acad. Sci. USA 98: 7504–7509
Esteller M, Corn PG, Baylin SB, Herman JG . 2001 Cancer Res. 61: 3225–3229
Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG . 1998 Cancer Res. 58: 4515–4518
Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG . 1999a Cancer Res. 59: 793–797
Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG . 1999b Cancer Res. 59: 67–70
Fruhwald MC, O'Dorisio MS, Dai Z, Rush LJ, Krahe R, Smiraglia DJ, Pietsch T, Elsea SH, Plass C . 2001a Genes Chrom. Cancer 30: 38–47
Fruhwald MC, O'Dorisio MS, Dai Z, Tanner SM, Balster DA, Gao X, Wright FA, Plass C . 2001b Oncogene 20: 5033–5042
Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM . 1997 J. Biol. Chem. 272: 22322–22329
Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB . 1996 Proc. Natl. Acad. Sci. USA 93: 9821–9826
Loeb LA . 2001 Cancer Res. 61: 3230–3239
Mares J, Kriz V, Weinhausel A, Vodickova S, Kodet R, Haas OA, Sedlacek Z, Goetz P . 2001 Cancer Lett. 166: 165–171
Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zöchbauer-Muller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A, Czerniak B, Gazdar AF . 2001 Cancer Res. 61: 8659–8663
Maruyama R, Toyooka S, Toyooka KO, Virmani AK, Zöchbauer-Muller S, Farinas AJ, Minna JD, McConnel J, Frenkel EP, Gazdar AF . 2002 Clin. Cancer Res. 8: 514–519
Mollerup S, Ryberg D, Hewer A, Phillips DH, Haugen A . 1999 Cancer Res. 59: 3317–3320
Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, Astuti D, Morgan NV, Moch H, Richards FM, Kishida T, Yao M, Schraml P, Latif F, Maher ER . 2001 Cancer Res. 61: 7277–7281
Nakamura M, Sugita K, Inukai T, Goi K, Iijima K, Tezuka T, Kojika S, Shiraishi K, Miyamoto N, Karakida N, Kagami K, Koyama T, Mori T, Nakazawa S . 1999 Leukemia 13: 884–890
Ohtani-Fujita N, Dryja TP, Rapaport JM, Fujita T, Matsumura S, Ozasa K, Watanabe Y, Hayashi K, Maeda K, Kinoshita S, Matsumura T, Ohnishi Y, Hotta Y, Takahashi R, Kato MV, Ishizaki K, Sasaki MS, Horsthemke B, Minoda K, Sakai T . 1997 Cancer Genet. Cytogenet. 98: 43–49
Sato M, Mori Y, Sakurada A, Fujimura S, Horii A . 1998 Hum. Genet. 103: 96–101
Takita J, Yang HW, Bessho F, Hanada R, Yamamoto K, Kidd V, Teitz T, Wei T, Hayashi Y . 2000 Med. Pediatr. Oncol. 35: 541–543
Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ . 2000 Nat. Med. 6: 529–535
Toyooka KO, Toyooka S, Virmani AK, Sathyanarayana UG, Euhus DM, Gilcrease M, Minna JD, Gazdar AF . 2001a Cancer Res. 61: 4556–4560
Toyooka S, Pass HI, Shivapurkar N, Fukuyama Y, Maruyama R, Toyooka KO, Gilcrease M, Farinas A, Minna JD, Gazdar AF . 2001b Cancer Res. 61: 5727–5730
Tsuchiya T, Tamura G, Sato K, Endoh Y, Sakata K, Jin Z, Motoyama T, Usuba O, Kimura W, Nishizuka S, Wilson KT, James SP, Yin J, Fleisher AS, Zou T, Silverberg SG, Kong D, Meltzer SJ . 2000 Oncogene 19: 3642–3646
Virmani AK, Rathi A, Zochbauer-Muller S, Sacchi N, Fukuyama Y, Bryant D, Maitra A, Heda S, Fong KM, Thunnissen F, Minna JD, Gazdar AF . 2000 J. Natl. Cancer Inst. 92: 1303–1307
Wong IH, Ng MH, Huang DP, Lee JC . 2000 Blood 95: 1942–1949
Yoon JH, Dammann R, Pfeifer GP . 2001 Int. J. Cancer 94: 212–217
Acknowledgements
This work was supported by grant U01CA8497102 from the Early Detection Research Network, National Cancer Institute, Bethesda, MD, USA.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Harada, K., Toyooka, S., Maitra, A. et al. Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene 21, 4345–4349 (2002). https://doi.org/10.1038/sj.onc.1205446
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1205446
Keywords
This article is cited by
-
DNA methylation in Hepatoblastoma-a literature review
Italian Journal of Pediatrics (2020)
-
Global DNA methylation profiling uncovers distinct methylation patterns of protocadherin alpha4 in metastatic and non-metastatic rhabdomyosarcoma
BMC Cancer (2016)
-
Epigenetic regulation of human retinoblastoma
Tumor Biology (2016)
-
The tumor suppressive role of RASSF1A in osteosarcoma through the Wnt signaling pathway
Tumor Biology (2016)
-
Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma
Modern Pathology (2015)