Abstract
Although the development of bone metastasis is a major detrimental event in prostate cancer, the molecular mechanisms responsible for bone homing and destruction remain largely unknown. Here we show that loss of miR-15 and miR-16 in cooperation with increased miR-21 expression promote prostate cancer spreading and bone lesions. This combination of microRNA endows bone-metastatic potential to prostate cancer cells. Concomitant loss of miR-15/miR-16 and gain of miR-21 aberrantly activate TGF-β and Hedgehog signaling, that mediate local invasion, distant bone marrow colonization and osteolysis by prostate cancer cells. These findings establish a new molecular circuitry for prostate cancer metastasis that was validated in patients' cohorts. Our data indicate a network of biomarkers and druggable pathways to improve patient treatment.
Similar content being viewed by others
Introduction
Skeletal metastases occur in more than 80% of cases of advanced-stage prostate cancer and are a major detrimental event for patients.1 Tumor cell homing into the bone marrow promotes dramatic alterations in osteoblast and osteoclast function and bone remodeling, causing severe lesions. Metastatic prostate cancer cells acquire a bone cell-like phenotype by a process, called osteomimicry, which allows their survival and proliferation in the bone marrow microenvironment. At the same time, osteoblasts and osteoclasts aberrantly modify their proliferation and differentiation programs during the metastatic process, thereby altering bone density.2, 3, 4 Although the different steps that characterize the development of prostate cancer metastases have been essentially defined, the molecular events that trigger and fuel the bone colonization and metastatic progression are still unknown.
Development of metastasis requires migratory and invasive capacity of tumor propagating cells to distant sites. Migration and invasion are features shared by undifferentiated tumor cells showing traits of epithelial–mesenchymal transition (EMT), a process driven by genetic and epigenetic signals greatly influenced by the tumor microenvironment. In the bone marrow microenvironment, stromal and metastatic prostate cancer cells produce TGF-β,5 which is released in sera of advanced patients.6 The ability of TGF-β inhibitors to prevent the formation of bone metastasis in preclinical models suggests that TGF-β may have a considerable role in prostate cancer progression.7 Aberrant TGF-β signaling can promote the expansion of tumor propagating cells and the conversion of early epithelial tumors to invasive and metastatic cancers through the induction of EMT.8 Another relevant pathway in prostate cancer may be represented by Hh signaling. Initial studies have shown that Hh targeting suppresses the growth of prostate cancer cell lines and displays therapeutic activity in prostate cancer xenografts.9, 10 More recent data indicate that IHH promotes the expansion of tumor propagating cells through the direct transcriptional control of the polycomb gene Bmi-1,11 which has a key role in prostate cancer initiation and progression.12
Compelling evidence demonstrates microRNAs (miRNAs) have a central role in controlling basic cell functions, including migration and invasion.13, 14 The expression of miRNAs is widely altered in cancer,15 suggesting that miRNA deregulations are deeply implicated in tumor development and progression.16, 17 Several articles demonstrate that miR-15 and miR-16 (miR-15/16) deregulation is associated with cancer progression by targeting several oncogenes, such as BCL2, CCND1, CCNE1 and CDK4-6, which promote cell proliferation and survival,18, 19, 20, 21 induction of BMI-122 and EMT through the regulation of AP4.23 Moreover, other data suggest that miR-15 and miR-16 control the expression of FGF-2, FGFR1, VEGF and WNT signaling,24, 25 which are able to promote tumor angiogenesis and bone metastases.
miR-21 is one of the most commonly and highly upregulated oncomir in many types of cancer.26 In prostate cancer, miR-21 promotes hormone-dependent and hormone-independent growth.27 Importantly, RAS induces the upregulation of miR-2128 and significantly contribute to the aggressiveness of several tumor types.29 Although not frequently mutated in prostate cancer, RAS isoforms have a pivotal role in multiple pathways that have been implicated in prostate tumorigenesis. Furthermore, RAS has been shown to promote prostate cancer progression by working synergistically with other pathways. In particular, a large body of literature indicates cooperation between RAS and TGF-β, including a prominent role of RAS signaling in the conversion from anti- to pro-oncogenic TGF-β signaling.30 Interestingly, the TGF-β inhibitor SMAD7 has been described as a target of the RAS downstream effector miR-21.31
The complex molecular events that occur at the level of miRNA and oncogene families during prostate metastasis formation are still unclear. Herein, we reveal a central role of the combined miR-21 upregulation and miR-15/miR-16 downregulation in the spread of metastases and in bone lesion formation. We clarify how RAS, TGF-β and Hedgehog signaling cooperate in concert with miR deregulation to promote cancer progression and bone alteration in prostate cancer.
Results
MiR-15 and MiR-16 Downregulation Promotes Tumor Growth and Invasion
We previously demonstrated that loss of miR-15 and miR-16 in epithelial prostate compartment has a pro-tumor effect in vitro and in vivo.19 In order to define the putative role of miR-15/miR-16 in metastatic spreading, we forced their downregulation in the early tumor cell line RWPE-2, which forms small subcutaneous tumors without having a metastatic effect in the NOD-scid IL2Rgamma-null (NSG) mouse model. For selectively sequestering both miRNAs, we used the lentiviral sponge vector TW3′Decoy15-16 (Decoy15-16) and TW3′ empty vector as relative control.19 RWPE-2 Decoy15-16 cells showed higher migratory and invasive capacity than control cells (Figure 1a). Such enhanced migration and invasion correlated with increased activation of AKT and ERK signaling19 and with overexpression of the prostate cancer marker Alpha-methylacyl-CoA racemase (Supplementary Figure 1a), which is associated with tumor expansion risk.32 The capacity of TW3’ and Decoy15-16 RWPE-2 cells to invade and colonize a tissue was then tested in vivo by injecting these cells into a permissive site, the subrenal capsule space of NSG mice. For tumor growth monitoring, cells were co-infected with a lentiviral vector encoding the luciferase reporter (TW-Luc) and analyzed in vivo by luminescence imaging (Supplementary Figure 1b). Three weeks after injection, RWPE-2 Decoy15-16 cells had doubled the expansion of control cells (Figure 1b). More importantly, RWPE-2 Decoy15-16 cells showed a pronounced invasion of the renal parenchyma coupled with new vessel formation, as indicated by hematoxylin and eosin (H&E) staining and CD31 immunofluorescence (Figure 1b). It is worthy to note that 9 weeks after injection, kidneys inoculated with TW3′ and Decoy15-16 RWPE-2 cells were both invaded by large tumor masses (Supplementary Figure 1c), but only Decoy15-16 cells were able to spread to distant organs, as indicated by the presence of lung metastasis (Figures 1c and d). Although the lung is an infrequent metastatic site for prostate cancer, in the subrenal capsule model it is the first organ colonisable by cells that enter the bloodstream. Thus, this assay demonstrated the capability of Decoy15-16 cells to enter blood flow and invade organs. Further, we orthotopically injected TW3’ and Decoy15-16 RWPE-2 cells into the anterior prostate of NSG mice (Figure 1e). Both the stereomicroscopy and the bioluminescence analyses at 5 weeks showed that Decoy15-16 cells grew extremely faster than the control population, suggesting that the loss of miR-15/miR-16 has a considerable effect in the context of the prostate microenvironment. Only Decoy15-16 cells were rarely able to colonize the liver after 15 weeks (Figure 1f). Thus, miR-15/miR-16 control organ-confined and distant invasion of prostate cancer cells.
MiR-15 and MiR-16 vector-mediated sequestering promotes metastatic spreading. (a) Boyden chamber assay performed on TW3′ and Decoy15-16 RWPE-2 cells using uncoated (left panel) or collagen-coated (right panel) membranes. Histograms report fold increase in migration/invasion of Decoy15-16 over TW3′ cells. Values are mean±s.d. of three independent experiments. The scale bar represents 100 μm. (b) TW3′ and Decoy15-16 RWPE-2 cells were injected in the subrenal capsule and evaluated by IVIS imaging system for luciferase detection after 3 weeks. Histograms report relative photon emission of Decoy15-16 RWPE-2-injected mice over control cells-injected mice. Lower left panels show transmitted light and EGFP images of TW3′ and Decoy15-16 RWPE-2-injected mice by stereomicroscopy. Lower right panels show H&E and CD31/DAPI staining of kidney sections. Nine mice for each group were used. Values are mean±s.d. The scale bar represent 3 mm. (c) Histogram represents the percentage of TW3′ and Decoy15-16 RWPE-2 injected mice with lung metastases after 9 weeks. One IVIS representative image of Decoy15-16 RWPE-2-injected mice was reported. Nine mice for each group were used. (d) Representative stereomicroscopic images and H&E stainings of Decoy15-16 RWPE-2-generated lung metastasis. The scale bar represent 3 mm. (e) TW3′ and Decoy15-16 RWPE-2 cells were injected into the mouse anterior prostate and evaluated by stereomicroscopy and IVIS imaging system after 9 weeks. Histogram showing relative photon emission of RWPE-2 Decoy15-16-injected mice over control TW3′ group. Ten mice for each group were used. Values are mean±s.d. The scale bar represents 3 mm. (f) Hepatic metastasis derived from Decoy15-16 RWPE-2 cells 15 weeks after injection into the murine anterior prostate. Histogram demonstrating the percentage of mice with liver metastasis. The scale bar represent 1 mm (Black) and 200 μm (white).
MiR-15 and MiR-16 downregulation causes bone colonization and lesions
The ability of prostate cancer cells to grow and metastasize in the bone suggests that these cells acquire an aberrant phenotype that promotes their growth and survival in the new microenvironment. To explore a potential difference in osteotropism, we injected TW3′ and Decoy15-16 RWPE-2 cells into the left ventricle of NSG mice to track their route by bioluminescence imaging. In contrast to the control population, Decoy15-16 cells engrafted with high frequency into mouse bones 3 weeks after injection, suggesting a critical role of miR-15 and miR-16 downregulation for bone marrow homing (Figure 2a). Moreover, to investigate whether miR-15/miR-16 downregulation confers to prostate cells the capability of growing in the bone microenvironment independently of an enhanced homing, we injected TW3’ and Decoy15-16 RWPE-2 cells co-infected with luciferase and β-galactosidase reporter genes directly in NSG mouse tibiae. Bioluminescence imaging and β-galactosidase staining 3 weeks after injection showed that RWPE-2 Decoy15-16 consistently engrafted in the bone, while RWPE-2 TW3′ control cells did not (Figures 2b and c), thus confirming the involvement of miR-15 and miR-16 loss in bone metastasis formation.
MiR-15 and MiR-16 deregulation are associated with bone lesions in prostate cancer. (a) Histogram shows the percentage of mice with bone bioluminescent signals 3 weeks after intracardiac injection with TW3′ and Decoy15-16 RWPE-2 cells. Representative images obtained by bioluminescence imaging. Ten mice for each group were used. (b) Histogram shows the percentage of mice with bone bioluminescent signals 3 weeks after injection of TW3′ and Decoy15-16 RWPE-2 cells into the tibial shaft. Representative images obtained by bioluminescence imaging. Twenty mice for each group were used. (c) Hematoxilyn/β-galactosidase (β-gal) staining of tibial shaft sections 3 weeks after injection with TW3′ and Decoy15-16 RWPE-2 cells. (d) MSCs cocultured with TW3’ and Decoy15-16 RWPE-2 cells in presence of mesenchymal stem cell medium, and evaluated for osteoblast differentiation by ALP staining. In the upper panels, ALP staining after 20 days of culture. In the lower panels, EGFP positivity and ALP staining after 10 days of co-culture. The scale bar represent 100 μm. On the right, histogram representing the percentage of EGFP positive cells in co-cultures, as determined by fluorescence activated cell sorting evaluation. Values are mean±s.d. of three independent experiments. (e) Histogram illustrating the percentage of EGFP positive cells in co-culture with osteoblasts (osteo) obtained by MSCs kept in osteoblast differentiation medium. Percentages were determined by fluorescence activated cell sorting evaluation. Values are mean±s.d. of three independent experiments. (f) Computed tomography analysis of tibiae injected with TW3′ and Decoy15-16 RWPE-2 cells 9 weeks after treatment. Single sections and radiographic images are reported. Ten mice for each group were used. (g) Evaluation of osteolysis (upper graph) and osteosclerosis (lower graph) in mice reported in (f). Osteolysis is calculated as percentage of empty spaces over total bone volume. Osteosclerosis is shown by relative increase in the bone volume of tibial shafts injected with TW3’ and Decoy15-16 RWPE-2 cells. Both graphs were obtained through the elaboration of morphometric parameters. (h) Representative H&E staining images of Decoy15-16 RWPE-2-injected tibiae after 9 weeks, revealing abnormal bone density and areas of lysis.
In order to evaluate the capability of RWPE-2 cells to influence bone renewal in vitro, TW3′ and Decoy15-16 RWPE-2 were co-cultivated with mesenchymal stem cells (MSCs)33, 34 which are the precursors of osteoblasts. Bone metastases in prostate cancer are typically osteoblastic and in the course of the metastatic process tumor stimuli induce MSCs to aberrantly differentiate, thereby causing osteosclerosis. Staining of the osteoblast marker alkaline phosphatase (ALP) was performed after 10 and 20 days of co-culture. Both TW3’ and Decoy15-16 RWPE-2 cells were able to induce differentiation of MSCs into osteoblasts in a medium for MSCs cultivation (Figure 2d). However, Decoy15-16 cells displayed a consistent proliferative advantage over control cells in co-culture conditions, both in medium for MSCs and in medium for osteoblast differentiation (Figures 2d and e), whereas MSC viability and proliferation were not affected (data not shown). Consistently with engraftment in mouse bones, radiographic and microtomographic analysis of mice injected with TW3’ or Decoy15-16 RWPE-2 cells showed that only Decoy15-16 RWPE-2 cells caused bone alterations, as revealed by detecting increased bone density as well as osteolysis (Figure 2f). The bone mineralization analysis confirmed a mixed osteosclerotic and osteolytic alteration (Figure 2g). H&E staining of tibiae injected with RWPE-2 Decoy15-16 confirmed the diffused mixed bone neo-formation and osteolysis areas (Figures 2g and h). These data suggest that miR-15/16 control the activity of prostate cancer key genes required for bone homing and lesions.
In order to explore the prognostic significance of miR-15 and miR-16 downregulation, we analyzed a publicly available data set for miRNA expression by using bioinformatic tools (NCBI GEO accession GSE21032).35 We included in the analysis 160 patients with documented clinical data and disease follow-up. The results showed that low expression of miR-15 and miR-16 is associated to bone metastasis and faster progression (Figures 3a and b).
MiR-15 and MiR-16 Downregulation is correlated with metastasis and poor prognosis. The prognostic impact of miR-15a and miR-16 expression was evaluated by the analysis of Taylor data set, which contains data from 99 primary tumors and 9 visceral and 5 bone metastases of prostate cancer patients. (a) Comparison of miR-15a and miR-16 expression in the 99 primary and 14 metastatic prostate tumors. (b) Comparison of miR-15a and miR-16 expression in the 99 primary tumors, 9 visceral and 5 bone metastases. (c) Kaplan–Meier survival analysis of disease-free survival of 35 patients with low miR-15a and miR-16 levels vs 62 patients without decreased miR-15 and miR-16 expression.
MiR-15 and MiR-16 Downregulation leads to hedgehog signaling activation
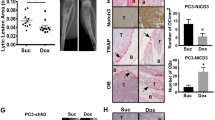
Although several known targets of miR-15a and miR-16 are involved in cancer homing and metastatic spreading, we searched for additional putative targets that may contribute to the development of prostate cancer metastasis. Using Targetscan software analysis, we found among possible candidates IHH, FGF-9, FGF18, PTHLH and CASR. Using the Taylor’s large gene data set, we first evaluated the mRNA level of these genes in primary and metastatic cancer tissues. All targets seemed to be expressed in primary tumors, but only IHH and PTHLH were significantly upregulated in bone metastases (Supplementary Figure 2a). When we examined the relationship between the expression pattern of these five genes and miR-15a/miR-16 levels, we found a significant inverse correlation (Figure 4a and data not shown). We then evaluated which of these genes were a direct target. Luciferase assay showed that only IHH was a direct target of miR-15 and miR-16 (Supplementary Figures 2b and c), while the others four genes were either not directly regulated or not correlated with metastasis formation, as from Taylor data set evaluation. The analysis of IHH mRNA level in several prostate cell lines showed that IHH is significantly upregulated in cell lines derived from metastatic tumors (Figure 4b). Furthermore, RWPE-2 cells dramatically increased IHH expression after lentiviral delivery of Decoy15-16 (Figures 4c and d), while the metastatic cell line PC3 considerably reduced IHH expression after miR-15 and miR-16 transduction (Figure 4c). Of note, BMI-1 is regulated by IHH at transcriptional level and resulted as a direct target of miR-15 and miR-16 in ovary cancer.22 Immunofluorescence analysis showed a consistent upregulation of BMI-1 in RWPE-2 Decoy15-16 cells (Supplementary Figure 2e), which could be mediated by both direct derepression of miR-15/miR-16 and transcriptional activation by IHH. In order to demonstrate a functional role of IHH in prostate cancer progression, we next silenced this gene using a mix of specific short hairpin RNAs in RWPE-2 Decoy15-16 cells. RWPE-2 Decoy15-16 cells transduced with specific short hairpin RNAs showed reduced capacity of cross-talk with microenvironment in vitro and in vivo as revealed by co-cultures with MSCs and by impaired homing when injected into tibial shaft of mice (Supplementary Figures 2e and f). By analyzing gene expression in patient cohorts, IHH expression was associated to recurrence and worse prognosis (Figures 4e and f and Supplementary Figure 2g).
Putative gene targets associated with metastases. (a) miR-16 expression level correlated with several putative gene targets and evaluated in 138 tumor tissues by analyzing Taylor tissue data set. Similar results were obtained with miR-15 (data not shown). (b) IHH mRNA level in non-neoplastic and prostate cancer cell lines and (c) in RWPE-2 UTR and Decoy15-16 and PC3 transduced with miR-15/16 and control vector, TW (*P<0.05; **P<0.01; ***P<0.001). (d) IHH protein level in RWPE-2 UTR and Decoy15-16 and evaluated by ELISA assay. (***P<0.001). (e) IHH mRNA relative expression in tissues of 104 patients free of recurrence (disease-free) and 27 showing recurrence by gene data set and clinical information analysis. (f) Evaluation of disease-free survival in 100 patients showing high and low level of IHH expression (50 patients for each group were analyzed).
Downregulation of MiR-15 and MiR-16 Leads to Hedgehog Signaling Activation through TGF-β enhanced activity
The biological effect of the loss of miR-15 and miR-16 is strictly dependent on the transcriptional activation of the relevant target genes, which should regain the expression and pre-existing molecular circuitry upon derepression. Several lines of evidence report that TGF-β is indispensable for bone maintenance and formation; this activity is mediated by RUNX-2 and RANKL genes.36, 37, 38, 39 Furthermore, the direct control of RUNX2 on IHH transcription and the feedback loop regulation of IHH on RUNX2 activity are well documented. We hypothesized a molecular circuitry involving RUNX2 and IHH at mRNA level. Therefore, we analyzed again the Taylor data set and found a direct correlation between RUNX2 and IHH levels in prostate cancer (Supplementary Figure 2e). Given the central role of TGF-β/IHH axis in the induction of bone metastases, we evaluated TGF-β signaling activation by monitoring phosphorylation of its downstream effector, the SMAD2/3 complex (pSMAD3). Western blotting analysis showed a significant increase of SMAD3 phosphorylation in RWPE-2 Decoy15-16 cells when compared with control cells, while TGF-β stimulation further enhanced the activation of SMAD signaling in Decoy15-16 cells (Supplementary Figure 3a). The levels of SMAD4, which forms a complex with SMAD2/3 for signal transduction to the nucleus, were also increased in Decoy15-16 cells (Supplementary Figure 3b). We next sought to determine how miR-15/16 control Smad4 level and Smad3 activity. Targetscan-driven bioinformatic analysis indicated USP9X as a potential target gene of miR-15 and miR-16. USP9X is an enzyme that has been reported to empower TGF-β signaling by deubiquitinating SMAD4, thereby facilitating its association with SMAD2/3.40, 41 We validated by the luciferase assay a direct binding of the two miRs to USP9X 3′UTR (Supplementary Figure 3c), thus confirming this gene as a new target of miR-15 and miR-16. Interestingly, in Xenopus embryos miR-15 and miR-16 have been reported to target activin RIIA, a receptor that can be triggered by activin A and Nodal, two ligands belonging to the TGF-β superfamily.42, 43 This targeting was confirmed in RWPE-2 Decoy15-16 cells (Supplementary Figure 3d). Thus, loss of miR-15 and miR-16 leads to derepression of TGF-β signaling by multiple routes. To investigate the role of Smad signaling in metastasis formation after miR-15/16 inactivation, we knocked down Smad4 in RWPE-2 Decoy15-16 cells. We first validated the interference capacity of a pool of SMAD4 siRNAs (Supplementary Figure 3e) and demonstrated by AnnexinV/7-AAD staining that this knockdown did not affect the viability of RWPE-2 Decoy15-16 cells in vitro (Supplementary Figure 3f). However, when injected into the tibia, SMAD4 siRNAs-transfected cells demonstrated a reduced engraftment as compared with scramble siRNA-transfected RWPE-2 Decoy15-16 (Supplementary Figure 3g), suggesting that in these experimental conditions the loss of miR-15/miR-16 increases TGF-β signaling and that this is causal for prostate cancer cell survival in bone marrow through gene activation cascade correlated and in synergy with IHH signaling. Consistently, RWPE-2 Decoy15-16 cells showed an increase of RUNX2, RANKL and additional genes associated with TGF-β signaling activation such as CTGF, ET-1, IL-11, IL-6, CXCR4, FGF-2 and BMP-4 mRNA (Supplementary Figure 3h).44 The TGF-β bone mediator gene, RANKL resulted correlated with bone metastases in the Taylor data set, and its expression was positively correlated with IHH level (Supplementary Figures 3i and l).
RAS Cooperates with MiR-15 and MiR-16 Downregulation for prostate cancer progression
We previously demonstrated that downregulation of miR-15 and miR-16 in the non-transformed prostate cell line RWPE-1 (RWPE-1 Decoy15-16) enabled these cells to grow in immunodeficient mice by enhancing their proliferation and invasion in vitro and in vivo.19 However, when injected in the left ventricle, luciferase-expressing RWPE-1 Decoy15-16 cells did not show bone signals after 5 weeks, while rare positive mice were countable 18 weeks after injection (data not shown). Since RWPE-2 derive from RWPE-1 cells by stable transformation with activated KRAS but are not per se metastatic, we hypothesized that RAS activation in RWPE-2 cells might synergize with miR-15 and miR-16 downregulation to promote bone colonization and subsequent lesions. Phosphorylation of the RAS effectors ERK1/2 in primary prostate tumor cells showed a consistent activation of the pathway, as compared with normal prostate cells (Supplementary Figure 4a). RAS aberrant activity has been reported to induce miR-21 in rat thyroid cells.28, 45 Moreover, miR-21 has been reported to be a major actor in RAS-mediated lung tumor progression since it sustains the signaling by repressing RAS inhibitors.29 Additionally, miR-21 levels have been associated with biochemical recurrence and metastatic hormone-refractory prostate cancer.46 Consistently, ERK phosphorylation correlated with miR-21 levels (Supplementary Figure 4b) and RWPE-2 cells express miR-21 about fourfold more than RWPE-1 cells (Supplementary Figure 4c). To further demonstrate the direct link between RAS and miR-21 in prostate cells, we interfered with RAS mRNAs using siRNAs for ki-RAS, H-RAS and N-RAS (Supplementary Figure 4d). All these siRNAs reduced miR-21 levels in RWPE-2 cells (Supplementary Figure 4e). As recently reported, miR-21 overexpression in non-tumorigenic RWPE-1 cells contribute to cell transformation by inducing expression of luminal markers and EMT.47 Although RWPE-1 miR-21 cells were unable to grow subcutaneously, contrary to the empty vector-transduced population, these cells engrafted and grew significantly when injected in the anterior prostate of the mouse (Supplementary Figure 5a), suggesting that miR-21 concurs in tumor development, but is insufficient to drive neoplastic transformation without other major lesions. Notably, miR-21 expression and maturation is enhanced by TGF-β signaling, while miR-21 itself has been reported to sustain TGF-β signaling by repressing its inhibitor SMAD7 in lung fibrosis.31 We confirmed and validated SMAD7 as a target of miR-21 in prostate cells (Supplementary Figure 5b). Thus, we hypothesized that increase in miR-21 and loss of miR-15/miR-16 might support each other in promoting TGF-β signaling activation. Of note, we observed that about 60% of primary prostate tumors with miR-15 and miR-16 downregulation displayed miR-21 levels higher than 1.5 over reference, indicating a frequent coexistence of these alterations (Supplementary Figure 5c). In order to demonstrate the specific role of miR-15 and miR-16 downregulation and miR-21 upregulation in promoting tumor engraftment and growth in vivo, we blunted the effect of aberrant expression of these miRNA in RWPE-2 Decoy15-16 cells using a lentiviral vector over expressing miR-15/16 and a Decoy21. RWPE-2 Decoy15-16 cells were infected with control vector (Control), with the vector containing miR-15 and miR-16 genes (miR-15/16) or Decoy21 (Decoy21). Cells were inoculated intratibially in NSG mice, which were analyzed 3 weeks after injection. As expected, control cells produced bone signals at high photon emission level in about 60% of mice, while restoring miR-15/16 levels essentially abolished the ability to engraft and grow into the bone promoted by the decoy vector (Supplementary Figure 5d). On the other hand, miR-21 suppression through Decoy21 vector did not alter the engraftment potential of RWPE-2 Decoy15-16 cells, but impaired consistently tumor growth and viability as showed by the reduction of photon emission levels (Supplementary Figure 5d). Thus, both loss of miR-15/16 and increase in miR-21 appear directly involved in bone lesion formation.
We then evaluated miR-21 level in tumors, correlating miR-21 levels with risk assessment parameters. The levels of miR-21 significantly increased in the higher Gleason scores (Figure 5a). Interestingly, we found high miR-21 level in T2 tumors of patients who showed recurrence (Figure 5b). Furthermore, we stratified patients for miR-15, miR-16 and miR-21 levels and found poorer prognosis and RUNX2/RANKL overexpression in patients who simultaneously showed miR-15/ miR-16 low and miR-21 high levels (Figures 5c and d). To determine whether this miRNA pattern correlates with advanced stages in prostate cancer patients, we microdissected 15 formalin-fixed and paraffin embedded prostate cancer tissues and 22 prostate tumor metastases (20 bone and 2 lymph node metastases), and analyzed miR-15/16 and miR-21 expression by real time PCR in purified prostate cancer cells. As reference and internal control, we used microdissected epithelial cells from a pool of three formalin-fixed normal prostate tissues. Among 15 patients, seven have been selected for 10 years negative follow-up monitoring and eight for clinical evidence of bone metastasis progression. MiR-15/miR-16 and miR-21 resulted simultaneously deregulated in 37.5% of primary tumors from patients with progression, in none of the patients with negative follow-up, and in 40.9% of patients with distant lesions (Supplementary Table S1), suggesting that this miRNA pattern may be a negative prognostic marker for prostate cancer. These results strongly support the hypothesis that miR-15 and miR-16 in cooperation with miR-21 may play a considerable role in prostate cancer progression and may represent good biomarkers for monitoring of cancer aggressiveness.
miR-21 cooperates in cancer progression. The prognostic impact of miR-21 expression was evaluated by the analysis of Taylor data set. (a) miR-21 expression in prostate cancer tumors with different Gleason scores. (b) Evaluation of miR-21 levels in prostate cancer tumors originally diagnosed T2 and selected for disease-free (N=60) or recurrence (N=9). (c) Evaluation of disease-free survival in patients showing four different patterns based on the level of miR-21 and miR-15a/miR-16. (d) RANKL and RUNX2 mRNA expression analyzed in tumors with different expression levels of miR-21 and miR-15a/miR-16.
In vitro synergistic activity of increase in MiR-21 and decrease in MiR-15/MiR-16
In order to verify a possible synergism between miR-21 upregulation and miR-15/miR-16 downregulation in TGF-β signaling activation and prostate cancer progression, we transduced Decoy15-16 RWPE-1 cells with miR-21 expressing vector, thus generating Decoy15-16/miR-21 RWPE-1 cells. We compared the phenotype and biological behavior of TW, miR-21, Decoy15-16 and Decoy15-16/miR-21 RWPE-1 cells. Phalloidin-mediated staining revealed a dramatic cytoskeleton remodeling in cells whose miRs were deregulated, with a massive formation of filopodia and lamellipodia and the acquisition of an elongated, fibroblast-like phenotype which was mostly pronounced in Decoy15-16/miR-21 cells (Supplementary Figure 6a). Such a phenotype is associated with EMT, a trans-differentiation process that anticipates metastatic spreading. As expected, miR-21, Decoy15-16 and Decoy15-16/miR-21 RWPE-1 cells showed an increase in vimentin and a decrease in E-cadherin expression, when compared with the control population (Supplementary Figures 7a and b). Consistently, miR-21 and Decoy15-16 RWPE-1 cells demonstrated increased invasion in collagen as compared with the control population, and again this feature was further enhanced in cells expressing both lesions (Supplementary Figure 7c). In addition, while RWPE-1 cells transduced with control vector were unable to grow in anchorage-independent conditions and RWPE-1 miR-21 only formed small clusters, the capability of RWPE-1 Decoy15-16 cells to grow in soft agar was enhanced in Decoy15-16/miR-21 cells, which were able to form very large colonies (Supplementary Figure 7d). During cancer progression, tumor cells gradually lose contact inhibition, programmed cell death response, and the capacity to form organized structures. In tumors deriving from acinar tissues, these aberrant features can be tested in vitro by three-dimensional (3D) cultures.48 We maintained TW, miR-21, Decoy15-16 and Decoy15-16/miR-21 RWPE-1 cells in 3D Matrigel cultures for about 1 week, then spheroids were plated on slides, stained with DAPI, and evaluated by confocal microscopy. In line with the normal developmental pathway, RWPE-1 TW cells created acini with a hollow lumen that results from the apoptosis of central cells. In contrast, miR-21 and Decoy15-16 RWPE-1 cells formed completely filled spheroid structures surrounded by spreading cells (Supplementary Figures 6b and c and Supplementary Video). Thus, miR-21 and Decoy15-16 RWPE-1 cells display invasive capacity coupled with apoptosis resistance and loss of polarity. Noteworthy, Decoy15-16/miR-21 RWPE-1 cells diffusely spread into the Matrigel and completely failed to form acinar structures (Supplementary Figure 6b). Interestingly, EMT has been associated with the cancer stem-like cells.49, 50, 51 The so called cancer stem-like cells express embryonic genes and are able to grow in serum-free and anchorage-independent conditions.52 By seeding our transduced populations in low-attachment culture plates using a medium optimized for cancer stem cell cultivation,53 we observed that, while TW, miR-21 and Decoy-15-16 RWPE-1 cells adhered to the plate and grew in areas with progressive larger size, Decoy15-16/miR-21 RWPE-1 cells formed foci and floating spheroidal structures (Supplementary Figure 6d), which closely resembled cancer stem cell cultures. Moreover, only the latter population was found positive for the embryonic gene SOX2 (Supplementary Figure 7e). Interestingly, SOX2 is under the transcriptional control of TGF-β, and its expression has been correlated with a more aggressive cancer phenotype.54 In a further effort to verify the synergism between decrease in miR-15/miR-16 and increase in miR-21, we demonstrated a considerable upregulation of the prostate cancer marker Alpha-methylacyl-CoA racemase in cells with both alterations (Supplementary Figure 6e) and, consistently with our initial hypothesis, we observed a strong activation of the TGF-β signaling in RWPE-1 Decoy15-16/miR-21 cells, as documented by SMAD3 phosphorylation (Supplementary Figure 6f). The targeting of USP9X was confirmed in RWPE-1 cells with miR-15/miR-16 downregulation (Supplementary Figure 6g). We then extended to these populations the expression analysis of the genes previously evaluated on RWPE-2 cells, which are associated with bone homing, osteomimicry, osteolysis and osteosclerosis. We observed a progressive increase of these mRNAs, with an overt overexpression in RWPE-1 Decoy15-16/miR-21 cells (Supplementary Figure 6h). Altogether, our data demonstrate that miR-15/miR-16 downregulation and miR-21 upregulation cooperate in TGF-β signaling activation and prostate cancer progression.
Raise of miR-21 and decrease of miR-15/16 are synergic at conferring metastatic properties
In order to test a putative synergistic effect of miR-15/miR-16 downregulation and miR-21 upregulation on organ-confined tumor spreading, TW, miR-21, Decoy15-16 and Decoy15-16/miR-21 RWPE-1 cells were injected under the renal capsule of NSG mice. After 3 weeks, some mice were sacrificed and kidney sections analyzed by immunofluorescence. Although TW and miR-21 populations remained localized in the site of injection, Decoy15-16 and especially Decoy15-16/miR-21 cells invaded the renal parenchyma and formed large lobular structures (Supplementary Figure 8a). Importantly, when analyzed 9 weeks after injection, we found that Decoy15-16/miR-21 RWPE-1 cells extravasated into the bloodstream and invaded lung lobes, where small microlesions were visible at high frequency, as documented by stereomicroscopic determination of EGFP positivity and by anti-EGFP and anti-HLA staining (Supplementary Figures 8b and e). Decoy15-16 RWPE-1 cells were rarely able to colonize the lung, forming small tumoral foci, whereas TW and miR-21 cells did not show any distant invasive capacity (Supplementary Figure 8c). Furthermore, we injected luciferase-expressing TW, miR-21, Decoy15-16 and Decoy15-16/miR-21 RWPE-1 cells in the left ventricle of NSG mice. Mice injected with Decoy15-16/miR-21RWPE-1 cells showed high frequency of bone signal as measured by luminescence imaging at 5 weeks post injection (Figure 6a). Moreover, since EGFP evaluation by flow cytometry is extremely sensitive, we evaluated the presence of EGFP positive cells into the bone marrow. As expected, only Decoy15-16/miR-21 RWPE-1 injected mice retained EGFP positive cells in the bone marrow (Figure 6b). Furthermore, an increasing number and intensity of lesions caused by Decoy15-16/miR-21 RWPE-1 cells was detected 18 weeks after injection (Figures 6c and d). Finally, Decoy15-16/miR-21 RWPE-1 cells injected directly into the tibial shaft revealed the capacity to engraft in a non-parental microenvironment. Both the radiographic and microtomographic analysis evaluation demonstrated osteosclerosis and osteolysis, indicating that these cells can cause bone lesions similarly to Decoy15-16 RWPE-2 cells (Figures 6e and f and Supplementary Figure 9). Thus, the considerable enhancement of TGF-β signaling induced by the modulation of miR-15/miR-16 and miR-21 promotes cancer progression and metastatic spreading.
MiR-15/miR-16 downregulation and miR-21 upregulation are sufficient to cause bone lesions in murine models. (a) TW, miR-21, Decoy15-16 and Decoy15-16/miR-21 RWPE-1 cells were intracardially injected in mice and evaluated by bioluminescence imaging. The histogram illustrates the percentage of mice with bone bioluminescent signals 5 weeks after the injection. Twelve mice for each group were used in three independent experiments. (b) Representative cytofluorimetric determination of EGFP positive cells among bone marrow cells obtained by phosphate-buffered saline flushing of tibiae and femurs of mice described in (a). The histogram represents the percentage of EGFP positive cells and is calculated as fold change over phosphate-buffered saline-injected mice (Ctr). Values are mean±s.d. of three independent experiments. (c) Representative bioluminescent images of whole mouse and dissected hind limb 18 weeks after intracardiac injection of RWPE-1 Decoy15-16/miR-21 cells. (d) Radiographic evaluation of whole mice intracardially injected with RWPE-1 Decoy15-16/miR-21. Phosphate-buffered saline-injected mice were used as control. (e) Representative image of bone luminescent signal after intratibial injection of RWPE-1 Decoy15-16 cells. Histogram represents the percentage of mice with bone bioluminescent signals 5 weeks after the injection. (f) Single section images obtained by immunohistochemistry and computed tomography analysis of tibiae injected with RWPE-1 Decoy15-16/miR-21 cells after 8 weeks. Nine mice for each group were used in three independent experiments.
Discussion
In this study we explored the molecular events causing bone lesions in murine models obtained with progressively transformed prostate cells. We demonstrated that miR-21 upregulation and loss of miR-15/miR-16 cluster cooperate to promote bone colonization and damage through the potentiation of TGF-β signaling. Such miRNA deregulation in prostate cancer promotes a plethora of effects mediated by the altered expression of their multiple targets. Aberrant TGF-β signal cascade can synergize with RAS, WNT, Hedgehog signaling, androgen receptor, FGF-2/FGFR1 axis, AKT and EGFR signaling in the context of an extremely aggressive phenotype that promote bone lesions.
Loss of MiR-15/MiR-16 cluster and upregulation of MiR-21 are critical events in the development of prostate cancer metastasis
We previously showed that advanced prostate cancers reduce miR-15/miR-16 cluster expression and that such a reduction was sufficient to transform non-neoplastic immortalized prostate cells.19 Since the loss of miR-15/miR-16 seems to be associated to advanced prostate cancer, we speculated that such a loss may be functionally correlated with bone metastases. Our data demonstrated that forcing miR-15 and miR-16 downregulation in RAS-transformed prostate cells considerably enhances both local invasion and metastatic activity. These cells are able to colonize and produce osteosclerosis and osteolysis into the bones of immunocompromised mice, indicating that the loss of miR-15/miR-16 in RAS-depending tumors is sufficient to cause bone lesions. RAS activation promotes the expression of miR-21, which in turn is associated in a positive feedback loop with RAS signaling. Our results confirmed that RAS activation increases miR-21 expression in prostate cancer cells. Furthermore, the analysis of non-transformed prostate cells engineered to express high levels of miR-21 and low levels of miR-15/miR-16 indicated that miR-21 mediates key effects of RAS on the development of bone metastasis. We observed a consistent increase in miR-21 in primary cells from more than half of patients with loss of miR-15/miR-16. Such concomitant alterations of miRNA levels may have detrimental effects in patients not undergoing rapid tumor debulking, as suggested by the striking correlation between this miRNA pattern and prostate cancer progression.
A number of target genes for miR-15/miR-16 and miR-21 have been discovered in solid tumors. These target genes control a variety of oncogenic cell functions, such as cell cycle, survival, migration and invasion.18, 55 The main features of prostate cancer cells with loss of miR-15/miR-16 and upregulation of miR-21 are increased aggressiveness, dedifferentiation and acquisition of a strong EMT phenotype, indicating that these alterations synergistically cooperate to maximize the malignancy of prostate cancer cells.
Enhancement of TGF-β signaling by deregulation of miR-15, miR-16 and miR-21
The cooperation between increased miR-21 and loss of miR-15/miR-16 in increasing prostate cancer malignancy seems to occur particularly at the level of TGF-β signaling. Notably, several targets of miR-15, miR-16 and miR-21 are related with the TGF-β pathway. Interestingly, miR-15 and miR-16 can target activin RIIA42, 43 a receptor belonging to the TGF-β family triggered by activin A and Nodal. The increased expression of Nodal reported in prostate cancer42, 56 may therefore contribute to enhance SMAD signaling after loss of miR-15 and miR-16. In addition, miR-21 expression and maturation is regulated by TGF-β family genes.31, 57 Herein, we show a new molecular circuitry driven by miR-15, miR-16 and miR-21 alterations and resulting in aberrant TGF-β signaling. TGF-β induces EMT, both in human non-neoplastic and tumor prostate cells, promoting the metastasis spreading. Our data further revealed a direct control of miR-15 and miR-16 on the SMAD4 deubiquitinase FAM (USP9X) and demonstrated that miR-21 targets SMAD7 in prostate cancer, thus contributing to enhance TGF-β signaling. SMAD7 transcription is controlled by TGF-β activation and can also inhibit activin and the bone morphogenetic proteins signaling,58 indicating a sophisticated feedback control of TGF-β and miR-21 regulation.
Published array-based screenings show a tumor gene signature associated with bone metastases.59, 60 Some of these critical genes are induced by TGF-β, which transactivates RANKL, RUNX2, CXCR-4, CTGF and IL-11, while indirectly promoting the sensitivity to bFGF cooperating in cancer progression.44 Furthermore TGF-β can post-transcriptionally regulate IHH ligand, while miR-15/miR-16 downregulation cause direct aberrant activation of both signaling, promoting aberrant bone remodeling and cancer progression. Our data showed that, upon miR-15/miR-16 loss and miR-21 upregulation, both non-neoplastic and early tumor cell lines acquired a bone-metastatic phenotype through the induction of genes associated to bone lesions. Thus, a number of TGF-β-related genes can be induced by deregulation of miR-15, miR-16 and miR-21 in prostate cancer. Although the contribution of each single gene remains to be identified, such pattern is consistent with a clear pro-metastatic activity due to the cooperation between TGF-β and these miRNA alterations (Supplementary Figure 10).
TGF-β-enhanced activity is associated with dedifferentiation and acquisition of a stem cell-like phenotype.61 A number of studies have shown the ability of Myc exogenous expression and embryonic genes to reprogram somatic cells.62 Interestingly, Myc overexpression downregulates miR-15 and miR-16,63 while miR-21 promotes EMT in prostate cancer.47 Such pattern of miRNA modulation is in line with our findings showing that miR-15 and miR-16 downregulation together with miR-21 upregulation induced a stem cell-like phenotype in prostate cancer, characterized by the capability to grow in anchorage-independent conditions, high clonogenicity and expression of embryonic genes such as SOX2. Interestingly, SOX2 has been described as a TGF-β target gene. Thus, the considerable enhancement of TGF-β signaling induced by the modulation of miR-15/miR-16 and miR-21 promotes a stem cell-like phenotype with features of EMT in prostate cancer cells and promotes cancer progression and metastatic spreading.
Clinical considerations and conclusions
Administration of targeted or conventional therapies requires accuracy of staging procedures and biomarkers predictive of patients’ response. Thus, although considerable efforts have been done in the attempt to identify patients at high risk of biochemical/radiological recurrence, currently available risk stratification models and predictive nomograms lack of the adequate accuracy. The proposed miRNA signature does not significantly correlate with higher Gleason scores or prostate specific antigen levels (data not shown), suggesting that this signature may add additional information over conventional analysis. The functional roles of miRNAs in tumor biology is deeply investigated and many evidences report that tissue or blood-based miRNA biomarkers that predict clinical behavior and/or therapeutic response can be used as prognostic and predictive biomarkers.64, 65 For the above reasons, our data suggest that miR-15 and miR-16 downregulation together with miR-21 upregulation should be investigated in order to verify whether these molecular parameters increase the accuracy of current predictors and, given the multiple molecular abnormalities related with their deregulation, as predictive biomarkers for optimal testing of innovative molecular targeted agents and bone-acting compounds. Furthermore, if validated in adequately powered clinical trials, our results can allow to optimize patients selection in clinical studies for bone metastasis prevention, which may envision the use of bone-acting agents in non-metastatic prostate cancer patients.66 Our results provide a rationale for biomarker-based bone metastasis prevention clinical trials, and define new ground for the development of novel therapies of advanced prostate cancer. Although our data may suggest new druggable pathways for bone metastases prevention and treatment, the therapeutic use of miRNA modulation in prostate cancer appears an extremely promising advancement toward more effective treatments.
Materials and methods
Animal models
Male 6–8-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Laboratory, Bar Harbor, ME, USA) were used for in vivo experiments. Luminescence-based whole animal imaging was obtained by IVIS 100 Imaging System (Caliper Life Sciences, Hopkinton, MA, USA).
For the subrenal capsule model, injection of 3 × 105 cells in 15 μl of Matrigel was administered with a 29G needle in the subcapsular space of the left kidney under a stereomicroscope. For the orthotopic prostate model, an injection of 3.5 × 105 cells in 15 μl of Matrigel was made in the left anterior prostate using a 29G needle under a stereomicroscope. For the intracardiac model, a percutaneous injection (1 × 105 cells in 50 μl of DPBS) was made in the left ventricle using a 29G needle. For the intratibial model, a small cavity was created in the tibial shaft of the right hind limb through the drilling of a 23G needle, and then 2.5 × 105 cells in 10 μl of Matrigel were inoculated using a 29G needle.
Cell culture and primary prostate cell isolation
RWPE-1 and RWPE-2 cells from ATCC were cultivated in supplemented keratinocyte serum-free medium and transduced with lentiviral particles produced by TW3′, TW3′Decoy15-16, TW, TW miR-21 vectors as previously described.19, 47 For RWPE-1 Decoy15-16/miR-21 generation, Decoy15-16 cells were infected with TW miR-21 and control vectors. TGF-β treatment was performed with 10 ng/ml for 24 h.
MSCs were isolated from human bone marrow and were cultivated in fresh alpha-medium (Invitrogen, Carlsbad, CA, USA) supplemented with 20% fetal calf serum (StemCell, Vancouver, BC, Canada). For osteogenic differentiation, MSCs were seeded in Osteogenesis Induction Medium (Cambrex, East Rutherford, NJ, USA) and cultured for 3 weeks. For co-culture experiments, cells were plated in a 1:1 ratio.
For three-dimensional cultures, cells suspended in culture medium were allowed to settle on Matrigel-coated multiwell plates; culture medium with 2% Matrigel was then added to each well. After 72 h microphotographs were taken. After 8 days spheroids were collected, allowed to settle on polylysine-coated coverslips, fixed, permeabilized and stained. Confocal Z-stack images were captured with a confocal microscope (Olympus FV1000, Tokyo, Japan) and used to generate three-dimensional reconstructions. Representative images of the equatorial (middle) plane were selected. For culture in 'stem cell' conditions cells were seeded in low-attachment plates in a medium optimized for cancer stem cell culture.
For prostate primary cells isolation, tissue samples were obtained from radical prostatectomies; tumor specimens and benign tissue from the prostate base were mechanically and enzymatically dissociated and the homogenate was put in culture in collagen-coated plates with BRFF-HPC1 medium (AthenaES, Baltimore, MD, USA).19
Formalin fixed and paraffin embedded tissues (metastases and primary tumors) were cut in 8 μm slides and arranged on laser-microdissection RNAse free slides (Nikon, Tokyo, Japan). They were maintained at 70 °C for 20 min, then treated with xilene for 20 min. Tissues were re-hydrated with scalar percentage of ethanol and distilled water and finally stained with Harris hematoxylin. Tumoral tissues were microdissected with a UV-laser microscope by Nikon. Laser cut samples were treated with TRIzol protocol for RNA extraction. Megaplex A retrotrascription and amplification system and miR-15/16, miR-21 and snU6RNA Taqman assay by Applied Biosystem (Carlsbad, CA, USA) were used for miRNA levels evaluation following recommended protocols.
Statistical analysis
Data are presented as the mean+s.d. of at least three or five experiments. Statistical significance of in vivo results was calculated by the t-test analysis and reported as±s.e.m. The immunofluorescence, western blot and fluorescence activated cell sorting results represent the three or five experiments. The data concerning microdissected tissues (Supplementary Table S1) were reported as mean of three independent experiments performed in duplicated.
Data set elaboration
miRNA, mRNA array data and clinical information from Taylor data set (NCBI GEO accession GSE21032) were accessed and explored through the MSKCC Prostate Cancer Genomics Data Portal (http://cbio.mskcc.org/prostate-portal/). Statistical analysis was performed by Student t-test or by one-way analysis of variance with Tukey post test. mRNA data are available for 29 normal prostates, 131 primary tumors (of which 27 with biochemical recurrence) and 19 metastatic samples (of which six localized to bones or spine). miRNA data are available for 28 normal prostates, 99 primary tumors (of which 19 with biochemical recurrence) and 14 metastatic samples (of which five localized to bones or spine). miR-15, miR-16 and miR-21 levels were compared with the mean value in the tumor samples group, which was set at 1. For miR-15/miR-16 Kaplan–Meier, only tumors with both miR-15 and miR-16 lower than the mean value were compared with all the others. For miR-15/miR-16/miR-21 Kaplan–Meier, tumors were assigned to the miR-15/miR-16<1 subset if the average expression of the two miRs was lower than 1. For IHH Kaplan–Meier, 50 patients with highest expression of the mRNA were compared with 50 patients with lowest expression. Kaplan–Meier survival curves were analyzed by Logrank P-test.
Accession codes
References
Sturge J, Caley MP, Waxman J . Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol 2011; 8: 357–368.
Mundy GR . Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002; 2: 584–593.
Weilbaecher KN, Guise TA, McCauley LK . Cancer to bone: a fatal attraction. Nat Rev Cancer 2011; 11: 411–425.
Logothetis CJ, Lin SH . Osteoblasts in prostate cancer metastasis to bone. Nat Rev Cancer 2005; 5: 21–28.
Teicher BA . Malignant cells, directors of the malignant process: role of transforming growth factor-beta. Cancer Metastasis Rev 2001; 20: 133–143.
Shariat SF, Kattan MW, Traxel E, Andrews B, Zhu K, Wheeler TM et al. Association of pre- and postoperative plasma levels of transforming growth factor beta(1) and interleukin 6 and its soluble receptor with prostate cancer progression. Clin Cancer Res 2004; 10: 1992–1999.
Mishra S, Tang Y, Wang L, deGraffenried L, Yeh IT, Werner S et al. Blockade of transforming growth factor-beta (TGFbeta) signaling inhibits osteoblastic tumorigenesis by a novel human prostate cancer cell line. Prostate 2011; 71: 1441–1454.
Massague J . TGFbeta in cancer. Cell 2008; 134: 215–230.
Sanchez P, Clement V, Ruiz i Altaba A . Therapeutic targeting of the Hedgehog-GLI pathway in prostate cancer. Cancer Res 2005; 65: 2990–2992.
Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci USA 2004; 101: 12561–12566.
Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 2006; 66: 6063–6071.
Lukacs RU, Memarzadeh S, Wu H, Witte ON . Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell 2010; 7: 682–693.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
Iorio MV, Croce CM . MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012; 4: 143–159.
Calin GA, Croce CM . MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866.
Esquela-Kerscher A, Slack FJ . Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269.
Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA . MicroRNAs – the micro steering wheel of tumour metastases. Nat Rev Cancer 2009; 9: 293–302.
Aqeilan RI, Calin GA, Croce CM . miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ 2010; 17: 215–220.
Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med 2008; 14: 1271–1277.
Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 2008; 36: 5391–5404.
Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010; 17: 28–40.
Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S et al. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res 2009; 69: 9090–9095.
Shi L, Jackstadt R, Siemens H, Li H, Kirchner T, Hermeking H . p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res 2014; 74: 532–542.
Musumeci M, Coppola V, Addario A, Patrizii M, Maugeri-Sacca M, Memeo L et al. Control of tumor and microenvironment cross-talk by miR-15a and miR-16 in prostate cancer. Oncogene 2011; 30: 4231–4242.
Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H . The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. Rna 2009; 15: 249–254.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257–2261.
Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res 2009; 69: 7165–7169.
Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M et al. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2009; 28: 73–84.
Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 2010; 18: 282–293.
Grusch M, Petz M, Metzner T, Ozturk D, Schneller D, Mikulits W . The crosstalk of RAS with the TGF-beta family during carcinoma progression and its implications for targeted cancer therapy. Curr Cancer Drug Targets 2010; 10: 849–857.
Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2010; 207: 1589–1597.
Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res 2002; 62: 2220–2226.
Sorrentino A, Ferracin M, Castelli G, Biffoni M, Tomaselli G, Baiocchi M et al. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Exp Hematol 2008; 36: 1035–1046.
Valtieri M, Sorrentino A . The mesenchymal stromal cell contribution to homeostasis. J Cell Physiol 2008; 217: 296–300.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22.
Das S, Tucker JA, Khullar S, Samant RS, Shevde LA . Hedgehog signaling in tumor cells facilitates osteoblast-enhanced osteolytic metastases. PLoS One 2012; 7 e34374.
Javelaud D, Pierrat MJ, Mauviel A . Crosstalk between TGF-beta and hedgehog signaling in cancer. FEBS Lett 2012; 586: 2016–2025.
Perrot CY, Javelaud D, Mauviel A . Overlapping activities of TGF-beta and Hedgehog signaling in cancer: therapeutic targets for cancer treatment. Pharmacol Ther 2013; 137: 183–199.
Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL et al. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev 2006; 25: 589–600.
Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 2009; 136: 123–135.
Stinchfield MJ, Takaesu NT, Quijano JC, Castillo AM, Tiusanen N, Shimmi O et al. Fat facets deubiquitylation of Medea/Smad4 modulates interpretation of a Dpp morphogen gradient. Development 2012; 139: 2721–2729.
Lawrence MG, Margaryan NV, Loessner D, Collins A, Kerr KM, Turner M et al. Reactivation of embryonic nodal signaling is associated with tumor progression and promotes the growth of prostate cancer cells. The Prostate 2011; 71: 1198–1209.
Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A et al. MicroRNA control of Nodal signalling. Nature 2007; 449: 183–188.
Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I et al. TGF-beta regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J 2011; 30: 783–795.
Frezzetti D, De Menna M, Zoppoli P, Guerra C, Ferraro A, Bello AM et al. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene 2011; 30: 275–286.
Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011; 71: 326–331.
Coppola V, Musumeci M, Patrizii M, Cannistraci A, Addario A, Maugeri-Sacca M et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene 2013; 32: 1843–1853.
Yamada KM, Cukierman E . Modeling tissue morphogenesis and cancer in 3D. Cell 2007; 130: 601–610.
Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011; 147: 759–772.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133: 704–715.
Polyak K, Weinberg RA . Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9: 265–273.
Clarke MF, Fuller M . Stem cells and cancer: two faces of eve. Cell 2006; 124: 1111–1115.
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007; 445: 111–115.
Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K . Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 2009; 5: 504–514.
Krichevsky AM, Gabriely G . miR-21: a small multi-faceted RNA. J Cell Mol Med 2009; 13: 39–53.
Vo BT, Khan SA . Expression of nodal and nodal receptors in prostate stem cells and prostate cancer cells: autocrine effects on cell proliferation and migration. Prostate 2011; 71: 1084–1096.
Davis BN, Hilyard AC, Lagna G, Hata A . SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008; 454: 56–61.
Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature 1997; 389: 631–635.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3: 537–549.
Kim C, Paik S . Gene-expression-based prognostic assays for breast cancer. Nat Rev Clin Oncol 2010; 7: 340–347.
Watabe T, Miyazono K . Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res 2009; 19: 103–115.
Takahashi K, Yamanaka S . Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676.
Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 2008; 40: 43–50.
Chan E, Prado DE, Weidhaas JB . Cancer microRNAs: from subtype profiling to predictors of response to therapy. Trends Mol Med 2011; 17: 235–243.
Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA . MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011; 8: 467–477.
Smith MR, Saad F, Coleman R, Shore N, Fizazi K, Tombal B et al. Denosumab and bone-metastasis-free survival in men with castration-resistant prostate cancer: results of a phase 3, randomised, placebo-controlled trial. Lancet 2012; 379: 39–46.
Acknowledgements
We thank Giuseppe Loreto, Tania Merlino, Laura De Salvo and Roberto Giaconella for their technical support. We thank Michelangelo Cordenonsi, Maria Musumeci and Marcello Maugeri-Saccà for manuscript discussion. This manuscript has been supported by National Ministry of Health, Under-forty researcher and Italy-USA microRNA program to DB and the Italian Association for Cancer Research (AIRC) and Fondazione Roma funding to RDM.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Bonci, D., Coppola, V., Patrizii, M. et al. A microRNA code for prostate cancer metastasis. Oncogene 35, 1180–1192 (2016). https://doi.org/10.1038/onc.2015.176
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.176
This article is cited by
-
Hsa-miR-15b-5p/miR-195-5p Controls Osteogenic Differentiation of Human Adipose-Derived Mesenchymal Stem Cells Through Regulating Indian Hedgehog Expression
Applied Biochemistry and Biotechnology (2023)
-
MiR-378a inhibits glucose metabolism by suppressing GLUT1 in prostate cancer
Oncogene (2022)
-
Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer
npj Genomic Medicine (2021)
-
miR-199b-5p-DDR1-ERK signalling axis suppresses prostate cancer metastasis via inhibiting epithelial-mesenchymal transition
British Journal of Cancer (2021)
-
MicroRNA-106a regulates autophagy-related cell death and EMT by targeting TP53INP1 in lung cancer with bone metastasis
Cell Death & Disease (2021)