Abstract
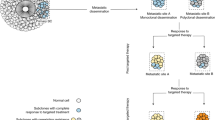
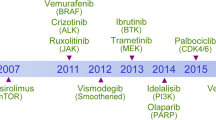
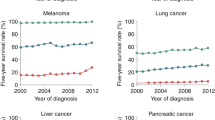
Molecular targeted therapy has the potential to dramatically improve survival in patients with cancer. However, complete and durable responses to targeted therapy are rare in individuals with advanced-stage solid cancers. Even the most effective targeted therapies generally do not induce a complete tumor response, resulting in residual disease and tumor progression that limits patient survival. We discuss the emerging need to more fully understand the molecular basis of residual disease as a prelude to designing therapeutic strategies to minimize or eliminate residual disease so that we can move from temporary to chronic control of disease, or a cure, for patients with advanced-stage solid cancers. Ultimately, we propose a shift from the current reactive paradigm of analyzing and treating acquired drug resistance to a pre-emptive paradigm of defining the mechanisms that result in residual disease, to target and limit this disease reservoir.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Kim Caesar/Nature Publishing Group

Kim Caesar/Nature Publishing Group
Similar content being viewed by others
References
Collins, F.S. & Varmus, H. A new initiative on precision medicine. N. Engl. J. Med. 372, 793–795 (2015).
Varmus, H. Ten years on—the human genome and medicine. N. Engl. J. Med. 362, 2028–2029 (2010).
Sawyers, C.L. The 2011 Gordon Wilson lecture: overcoming resistance to targeted cancer drugs. Trans. Am. Clin. Climatol. Assoc. 123, 114–123, discussion 123–125 (2012).
Sawyers, C.L. Lessons learned from the development of kinase inhibitors. Clin. Adv. Hematol. Oncol. 7, 588–589 (2009).
Sawyers, C.L. Shifting paradigms: the seeds of oncogene addiction. Nat. Med. 15, 1158–1161 (2009).
de Bono, J.S. & Ashworth, A. Translating cancer research into targeted therapeutics. Nature 467, 543–549 (2010).
Garraway, L.A. & Jänne, P.A. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2, 214–226 (2012).
Blumenthal, G.M. et al. Overall response rate, progression-free survival, and overall survival with targeted and standard therapies in advanced non-small-cell lung cancer: US Food and Drug Administration trial-level and patient-level analyses. J. Clin. Oncol. 33, 1008–1014 (2015).
Boxer, R.B., Jang, J.W., Sintasath, L. & Chodosh, L.A. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell 6, 577–586 (2004).
Zhang, Z. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 44, 852–860 (2012).
Hrustanovic, G. et al. RAS–MAPK dependence underlies a rational polytherapy strategy in EML4–ALK-positive lung cancer. Nat. Med. 21, 1038–1047 (2015).
Yu, H.A. et al. Analysis of tumor specimens at the time of acquired resistance to EGFR–TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 19, 2240–2247 (2013).
Van Allen, E.M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014).
Ahronian, L.G. et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 5, 358–367 (2015).
Poulikakos, P.I., Zhang, C., Bollag, G., Shokat, K.M. & Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signaling in cells with wild-type BRAF. Nature 464, 427–430 (2010).
Lito, P., Rosen, N. & Solit, D.B. Tumor adaptation and resistance to RAF inhibitors. Nat. Med. 19, 1401–1409 (2013).
Poulikakos, P.I. et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAFV600E. Nature 480, 387–390 (2011).
Lito, P. et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 (2012).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAFV600E inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Alizadeh, A.A. et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 21, 846–853 (2015).
de Bruin, E.C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Hiley, C., de Bruin, E.C., McGranahan, N. & Swanton, C. Deciphering intratumor heterogeneity and temporal acquisition of driver events to refine precision medicine. Genome Biol. 15, 453 (2014).
Burrell, R.A. & Swanton, C. Tumor heterogeneity and the evolution of polyclonal drug resistance. Mol. Oncol. 8, 1095–1111 (2014).
Turke, A.B. et al. Preexistence and clonal selection of MET amplification in EGFR-mutant NSCLC. Cancer Cell 17, 77–88 (2010).
Rosell, R. et al. Pretreatment EGFRT790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin. Cancer Res. 17, 1160–1168 (2011).
Lin, L. et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat. Genet. 47, 250–256 (2015).
Janku, F. et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One 6, e22769 (2011).
Hata, A.N. et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 22, 262–269 (2016).
Müller, J. et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 5, 5712 (2014).
Straussman, R. et al. Tumor microenvironment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012).
Blakely, C.M. et al. NF-κB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. 11, 98–110 (2015).
Sharma, S.V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Lee, H.J. et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26, 207–221 (2014).
Haq, R. et al. Oncogenic BRAF regulates oxidative metabolism via PGC-1α and MITF. Cancer Cell 23, 302–315 (2013).
Wilson, T.R. et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505–509 (2012).
Obenauf, A.C. et al. Therapy-induced tumor secretomes promote resistance and tumor progression. Nature 520, 368–372 (2015).
Bollag, G. et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599 (2010).
Szmulewitz, R.Z. & Ratain, M.J. Vemurafenib oral bioavailability: an insoluble problem. J. Clin. Pharmacol. 54, 375–377 (2014).
Undevia, S.D., Gomez-Abuin, G. & Ratain, M.J. Pharmacokinetic variability of anticancer agents. Nat. Rev. Cancer 5, 447–458 (2005).
Neesse, A., Algül, H., Tuveson, D.A. & Gress, T.M. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut 64, 1476–1484 (2015).
Jain, R.K. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell 26, 605–622 (2014).
Jänne, P.A. et al. Phase 1 safety and pharmacokinetic study of the PI3K–mTOR inhibitor SAR245409 (XL765) in combination with erlotinib in patients with advanced solid tumors. J. Thorac. Oncol. 9, 316–323 (2014).
Lin, N.U. Targeted therapies in brain metastases. Curr. Treat. Options Neurol. 16, 276 (2014).
Thompson, C.B. Targeting the anti-apoptotic signaling pathway. Clin. Adv. Hematol. Oncol. 7, 819–822 (2009).
Shaw, A.T. et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 370, 1189–1197 (2014).
Sequist, L.V. et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N. Engl. J. Med. 372, 1700–1709 (2015).
Jänne, P.A. et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N. Engl. J. Med. 372, 1689–1699 (2015).
Johnson, D.B. et al. Combined BRAF (dabrafenib) and MEK inhibition (trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J. Clin. Oncol. 32, 3697–3704 (2014).
Charakidis, M. & Boyer, M. Targeting MET and EGFR in NSCLC—what can we learn from the recently reported phase 3 trial of onartuzumab in combination with erlotinib in advanced non-small cell lung cancer? Transl. Lung Cancer Res. 3, 395–396 (2014).
Riely, G.J. et al. Prospective assessment of discontinuation and reinitiation of erlotinib or gefitinib in patients with acquired resistance to erlotinib or gefitinib followed by the addition of everolimus. Clin. Cancer Res. 13, 5150–5155 (2007).
Wagle, N. et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF–MEK inhibition. Cancer Discov. 4, 61–68 (2014).
Flaherty, K.T. et al. Combined BRAF and MEK inhibition in melanoma with BRAFV600 mutations. N. Engl. J. Med. 367, 1694–1703 (2012).
Long, G.V. et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N. Engl. J. Med. 371, 1877–1888 (2014).
Bhang, H.E. et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 21, 440–448 (2015).
Bozic, I. et al. Evolutionary dynamics of cancer in response to targeted combination therapy. eLife 2, e00747 (2013).
Schwaederle, M. et al. On the road to precision cancer medicine: analysis of genomic biomarker actionability in 439 patients. Mol. Cancer Ther. 14, 1488–1494 (2015).
Crystal, A.S. et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346, 1480–1486 (2014).
Gao, D. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 159, 176–187 (2014).
Gingras, I., Salgado, R. & Ignatiadis, M. Liquid biopsy: will it be the 'magic tool' for monitoring response of solid tumors to anticancer therapies? Curr. Opin. Oncol. 27, 560–567 (2015).
Li, X. et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 20, 769–777 (2014).
Karthaus, W.R. et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159, 163–175 (2014).
Piotrowska, Z. et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of EGFRT790-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 5, 713–722 (2015).
Nakasone, E.S. et al. Imaging tumor–stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell 21, 488–503 (2012).
Mayer, R.J. et al. Intensive postremission chemotherapy in adults with acute myeloid leukemia. N. Engl. J. Med. 331, 896–903 (1994).
Coombs, C.C., Tallman, M.S. & Levine, R.L. Molecular therapy for acute myeloid leukemia. Nat. Rev. Clin. Oncol. (2015).
Condeelis, J. & Weissleder, R. In vivo imaging in cancer. Cold Spring Harb. Perspect. Biol. 2, a003848 (2010).
Doebele, R.C. et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small-cell lung cancer. Clin. Cancer Res. 18, 1472–1482 (2012).
Van Allen, E.M. et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 20, 682–688 (2014).
De Silva, N. et al. Molecular effects of lapatinib in the treatment of HER2-overexpressing oesophago-gastric adenocarcinoma. Br. J. Cancer 113, 1305–1312 (2015).
Thress, K.S. et al. Acquired EGFRC797S mutation mediates resistance to AZD9291 in non-small-cell lung cancer harboring EGFRT790M. Nat. Med. 21, 560–562 (2015).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 795–801 (2015).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Behbehani, G.K. et al. Mass cytometric functional profiling of acute myeloid leukemia defines cell-cycle and immunophenotypic properties that correlate with known responses to therapy. Cancer Discov. 5, 988–1003 (2015).
Macosko, E.Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Larkin, J. et al. Combined nivolumab and ipilimumab, or monotherapy, in untreated melanoma. N. Engl. J. Med. 373, 23–34 (2015).
Sequist, L.V. et al. Randomized phase 2 study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J. Clin. Oncol. 29, 3307–3315 (2011).
Spigel, D.R. et al. Final efficacy results from OAM4558g, a randomized phase 2 study evaluating MetMAb or placebo in combination with erlotinib in advanced NSCLC (ASCO Annual Meeting) 7505 (American Society of Clinical Oncology, Alexandria, Virginia, USA, 2011).
Price, K.A. et al. Phase 2 trial of gefitinib and everolimus in advanced non-small-cell lung cancer. J. Thorac. Oncol. 5, 1623–1629 (2010).
Witta, S.E. et al. Randomized phase 2 trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J. Clin. Oncol. 30, 2248–2255 (2012).
Johnson, M.L. et al. Phase 1/2 study of HSP90 inhibitor AUY922 and erlotinib for EGFR-mutant lung cancer with acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors. J. Clin. Oncol. 33, 1666–1673 (2015).
Planchard, D. et al. Interim results of a phase 2 study of the BRAF inhibitor (BRAFi) dabrafenib (D) in combination with the MEK inhibitor trametinib (T) in patients (pts) with BRAFV600E mutated (mut) metastatic non-small-cell lung cancer (NSCLC) (ASCO Annual Meeting) 8006 (American Society of Clinical Oncology, Alexandria, Virginia, USA, 2011).
Yaeger, R. et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clin. Cancer Res. 21, 1313–1320 (2015).
Hurvitz, S.A. et al. Combination of everolimus with trastuzumab plus paclitaxel as first-line treatment for patients with HER2-positive advanced breast cancer (BOLERO-1): a phase 3, randomized, double-blind, multicenter trial. Lancet Oncol. 16, 816–829 (2015).
Swain, S.M. et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N. Engl. J. Med. 372, 724–734 (2015).
Acknowledgements
T.G.B. acknowledges funding support from the US National Institutes of Health Director's Office and Common Fund (New Innovators Award DP2-CA174497), the National Cancer Institute (R01-CA169338), the HHMI Collaborative Innovation Award Program, the Searle Scholars Program, the Pew Charitable Trust, and the Addario Lung Cancer Foundation. R.C.D. acknowledges funding support from the V Foundation for Cancer Research and the University of Colorado Lung Cancer SPORE.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
T.G.B. has ownership in Driver Group, is a consultant to Driver Group, Novartis, Astellas, Natera, Array Biopharma, and Ariad, and is a recipient of research grants from Servier and Ignyta. R.C.D. is a consultant to Array BioPharma and Ariad, and has received honoraria from AstraZeneca, Clovis, and Pfizer, research grants from Loxo Oncology, Mirati Therapeutics, and Abbott Molecular, and licensing fees from Chugai, Ariad, Blueprint Medicines, GVKbio, and Abbott Molecular.
Rights and permissions
About this article
Cite this article
Bivona, T., Doebele, R. A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nat Med 22, 472–478 (2016). https://doi.org/10.1038/nm.4091
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4091
This article is cited by
-
Mcl-1 mediates intrinsic resistance to RAF inhibitors in mutant BRAF papillary thyroid carcinoma
Cell Death Discovery (2024)
-
Improvement of ACK1-targeted therapy efficacy in lung adenocarcinoma using chloroquine or bafilomycin A1
Molecular Medicine (2023)
-
Transient targeting of BIM-dependent adaptive MCL1 preservation enhances tumor response to molecular therapeutics in non-small cell lung cancer
Cell Death & Differentiation (2023)
-
Durable responses to alectinib in murine models of EML4-ALK lung cancer requires adaptive immunity
npj Precision Oncology (2023)
-
Treatment of advanced non-small cell lung cancer with driver mutations: current applications and future directions
Frontiers of Medicine (2023)